Figures & data
Figure 1. Schematic representation of murine retinal explant culture and three different sample handling and embedding methods. (a) The neural retina was isolated from the enucleated mouse eye as a cup shape and four radial incisions made to flatten the retina into a flower shape. The retinal explant was cultured on a microporous membrane of a cell culture insert with the retinal ganglion cell (RGC) layer facing up. (b) The initial two approaches to sample processing involved three sample handling steps, including transfer to a 24-well plate for fixation, transfer to a hard plastic mesh cassette (Method #1) or biopsy cassette insert (Method #2) for processing, and positioning the whole explant along one petal edge for embedding. (c-i) The Histogel embedding method (Method #3) eliminated sample handling by fixing the explants on the culture inserts and transferring the entire insert onto a cold plate (d). Histogel was pipetted directly onto and encapsulated the explant (e). A weighing spatula was used to transfer the embedded explant onto a biopsy wrap, which was folded over and placed into a cassette (f). Following processing, the Histogel was clear and firm (g). A razor blade was used to bisect the explant so that each half contained central and peripheral retina (h). Each half was embedded on edge diagonally in an embedding mold (i).
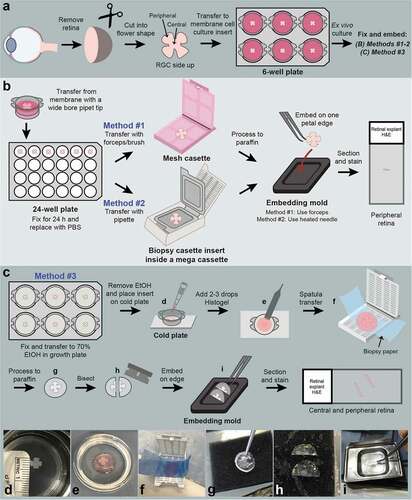
Figure 2. Histogel improves visualization of tissue in formalin-fixed paraffin embedded (FFPE) blocks. (a) Representative FFPE blocks of retinal explants embedded with and without Histogel. Histogel appears whiter than paraffin, enabling easy identification of tissue in the block (red arrows). (b) Improved visualization reduces the chance of sectioning past explant tissue.

Figure 3. Representative H&E-stained sections of retinal explants from each sample handling and embedding approach. (a-b) Explant. no culture, embedded with Method #3. Non-cultured retinal explants embedded with Method #3 demonstrated all layers of the neural retina with normal morphology and cellularity. (c-h) Cultured explants showed variable degeneration of retinal ganglion cells and photoreceptor processes as well as cell-cell dyshesion. (c) Sample embedded using Method #1 shows severe fragmentation and tangential sectioning (d) and frequent separation of retinal layers (black arrows) (e-f). Sample embedded using Method #2 shows smaller sections of peripheral retina with less fragmentation, improved orientation and layer continuity. Black arrows in (f) indicate focal tangential sectioning. (g-h) Samples embedded using Method #3 show minimal fragmentation, proper orientation, and improved representation of full-thickness retinal cross sections. INL, inner nuclear layer, ONL; outer nuclear layer; and RGC, retinal ganglion cell layer. Scale bar = 1000 μm (a, c, e, g); 100 μm (b, d, f, h).
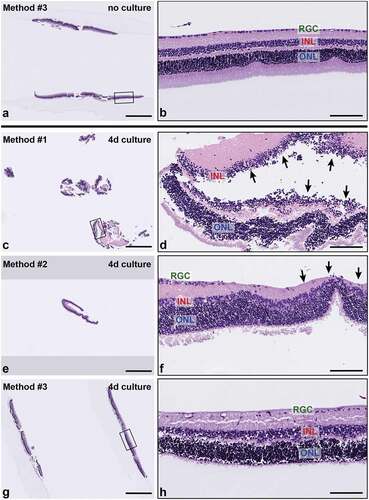
Figure 4. Histogel embedding improves multiple metrics of sample integrity and tissue quality. (a-d) H&E-stained sections of retinal explants from each embedding method were scored based on four metrics: lateral fragmentation (a), tissue orientation on edge (b), continuity of retinal layers (c), and the percentage of tissue with a full-thickness cross section (d). Within each embedding method, samples were divided into those not cultured or minimally cultured (‘-’) and those cultured ex vivo for 4 days (‘+’). Histogel-embedded samples show superior performance in the extent of sample fragmentation, sample orientation, and full thickness layer representation.
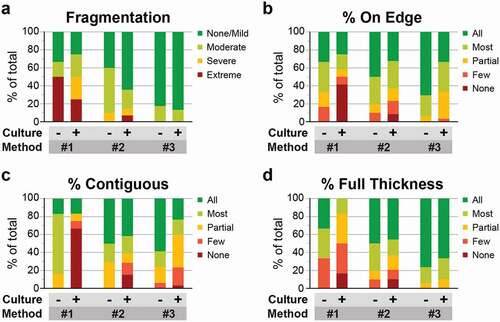
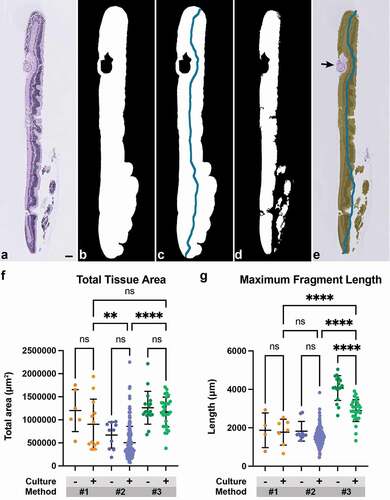
Figure 5. Use of Histogel increases the amount of analyzable tissue per slide. (a) Representative H&E-stained section of a retinal explant. (b) Tissue area was obtained by converting images to grayscale and Otsu thresholding to produce a binary mask. (c) Topological thinning and pruning were performed on this binary mask to obtain a filament representing tissue length for each fragment. (d) Whitespace was removed from both area and length measurements. (e) Additional artifacts, such as fragment of lens capsule (black arrow), were manually excluded from the captured area. (f) Total tissue area and (g) maximum fragment length were compared among minimally cultured or non-cultured (‘-’) and cultured (‘+’) samples across threeembedding methods. Method #3 Histogel-embedded cultured explants has significantly higher tissue area per slide than Method #2 samples and higher fragment length compared to Methods #1 and #2 (One-way ANOVA with post-hoc Šidak’s test for multiple comparisons; **, p < 0.01; ****, p < 0.0001). Scale bar = 100 μm.