Figures & data
Figure 1. Taxonomic boundaries in the Splendid Fairywren complex. (a) Distribution of -the Splendid Fairywren complex with colours indicating the range of the four currently recognised subspecies and approximate locations of hybrid zones between each of them—M. s. splendens (purple), M. s. callainus (turquoise), M. s. melanotus (dark blue), M. s. emmottorum (light blue). Localities of specimens examined in this study are indicated by black circles. Photos were obtained from the Macaulay Library at the Cornell Lab of Ornithology—M. s. splendens (ML611751385; Gosnells, Western Australia), M. s. callainus (ML519296141; Gawler Ranges, South Australia), M. s. melanotus (ML481245791; Mildura, Victoria), M. s. emmottorum (ML246987571; Barcoo, Queensland). (b) Population structuring of eight autosomal nuclear loci sampled across 36 Splendid Fairywrens with missing data for no more than 2 loci. Plots show results of STRUCTURE analyses for 2, 3, and 4 populations (K).
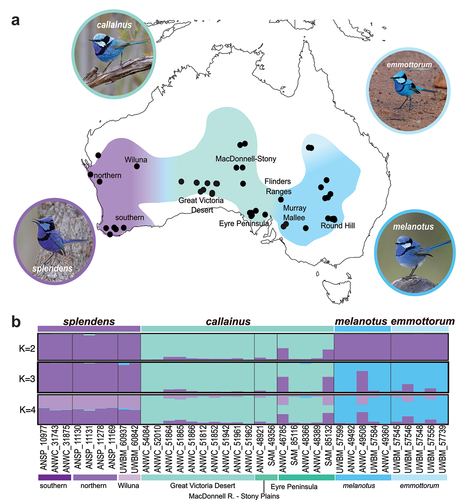
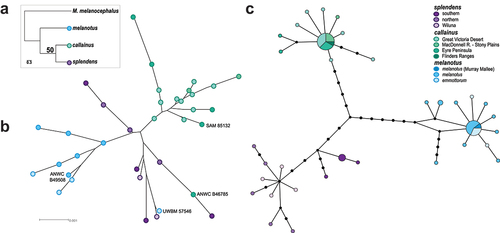
Figure 2. Nuclear and mtDNA variation across the Splendid Fairywren complex. (a) Species tree estimated from all concatenated data. (b) Unrooted Neighbor-joining tree estimated for the unphased concatenated 8-locus dataset. The museum accession number of specimens identified with admixed ancestry in STRUCTURE analyses are included for reference. (c) MtDNA ND2 haplotype network combined from specimens in the studies of Kearns et al. (Citation2009) and Dolman and Joseph (Citation2012).
Supplemental Material
Download PDF (2.5 MB)Data availability statement
Nuclear DNA sequences generated for this study are lodged with GenBank under the following accessions (PP412882 – PP413260). MtDNA sequences generated by previous studies can be found on GenBank under the following accessions (Kearns et al. (Citation2009): EU144234- EU144303 and EU534188-EU534192; Dolman and Joseph (Citation2012): JQ027448-JQ027433).
