Figures & data
Table 1. Summary of Vibrio vulnificus genomes used in the study.
Figure 1. (A) Heat-map of Euclidean distance matric calculated based on percentage values of average nucleotide identity (ANI) values of different strains of Vibrio vulnificus reveals two major groups. The strain sequenced in this study is highlighted in the red box. Details of genomes retrieved from NCBI GenBank are provided in . The values indicate the extent of dissimilarity from dark pink to green color; (B) MLST based phylogenetic analysis of V. vulnificus genomes and details of their country, source, and ST shows differential branching pattern based on vcg alleles. (vcg-C—red; vcg-E—green). the newly sequenced isolate is marked with ‘a star’; (C) Core genome-based phylogenetic analysis of V. vulnificus genomes showing two distinct lineages based on vcg alleles (vcg-C marked in red and vcg-C marked in green color).
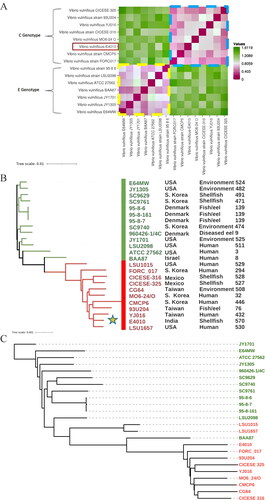
Table 2. Summary of the (A) assembly and annotation; (B) virulence genes identified by VFDB; (C) antimicrobial resistance genes identified by PATRIC in the genome of Vibrio vulnificus E4010 genome.
Figure 2. Schematic representation of virulence and fitness genes identified in Vibrio vulnificus E4010.
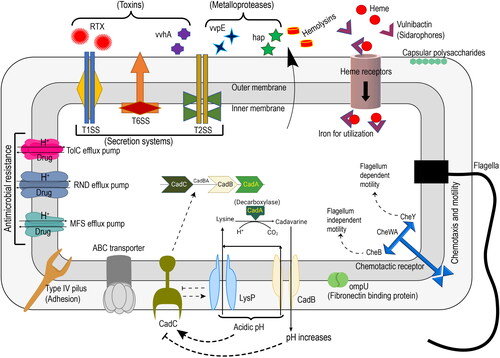
Figure 3. (A) BLAST atlas for chromosome 1 and 2 of Vibrio vulnificus E4010 generated using BRIG with CMCP6 as reference; (B) Schematic representation of Multifunctional autoprocessing repeats-in-toxin (MARTX) gene clusters in V. vulnificus.
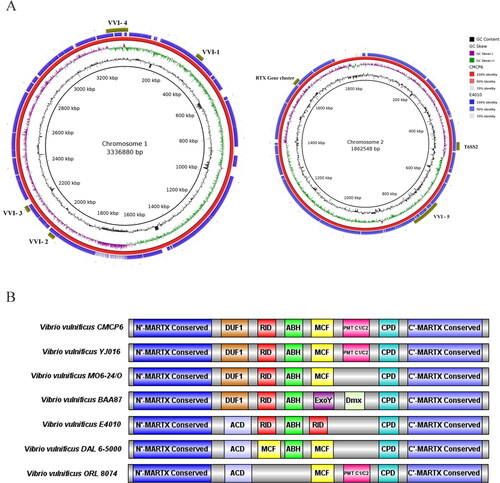
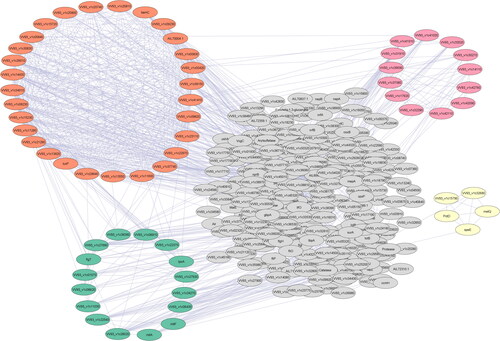
Figure 4. Basic PPI network of Vibrio vulnificus E4010 constructed using STRING consisting of 375 nodes and 1606 edges with clusters highlighted.
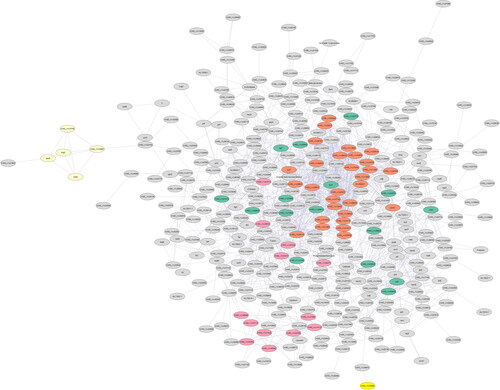
Figure 5. (A) the degree distribution shows a scale-free topology and measures up to a power law; (B) the average path length distribution (APL = 3.905) of the Vibrio vulnificus E4010 secretory network.
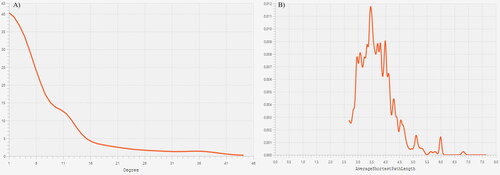
Table 3. The nodes with BC and/or degree values more than the mean plus standard deviation, and the majority of the proteins/nodes in the secretome network of Vibrio vulnificus E4010.
Supplemental Material
Download Zip (2.9 MB)Data availability statement
The draft genome of Vibrio vulnificus E4010 was earlier deposited in DDBJ/ENA/GenBank and given the accession WRPB00000000. The version described in this paper is version WRPB01000000. The new MLST profiles of V. vulnificus E4010 (pubMLST id: 775) have also been submitted to the pubMLST database (https://pubmlst.org).