Figures & data
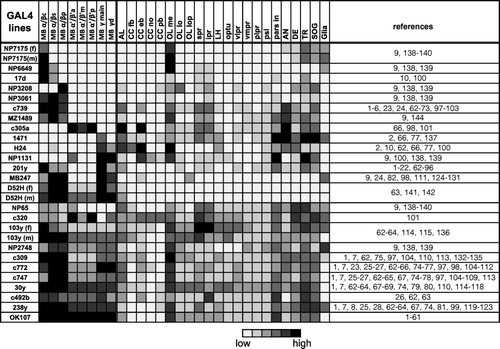
Figure 1 Expression pattern of 25 GAL4 lines. Summary of the expression levels of 25 MB-GAL4s in various brain areas defined by anti-Synapsin immunostaining. Gray scale indicates subjectively evaluated signal intensity. Note that a higher level of fluorescent signals in the certain brain area can result from larger population of GAL4 expressing cells and/or stronger GAL4 expression in each cell. MB, mushroom body; c, core subdivision: s, surface subdivision; p, posterior subdivision; a, anterior subdivision; m, middle subdivision; p, posterior subdivision; d, dorsal subdivision; AL, antennal lobe; CC, central complex; fb, fan-shaped body; eb, ellipsoid body; no, noduli; pb, protocerebral bridge; OL, optic lobe; me, medulla; lo, lobula; lop, lobula plate; spr, superior protocerebrum; ipr, inferior protocerebrum; LH, lateral horn; optu, optic tubercle; vlpr, ventrolateral protocerebrum; plpr, posteriorlateral protocerebrum; vmpr, ventromedial protocerebrum; psl, posterior slope; pars in, pars intercerebralis; AN, antennal nerve; DE, deutocerebrum; TR, tritocerebrum; SOG, subesophageal ganglion. References where these GAL4 drivers have been used are listed in Supplementary .
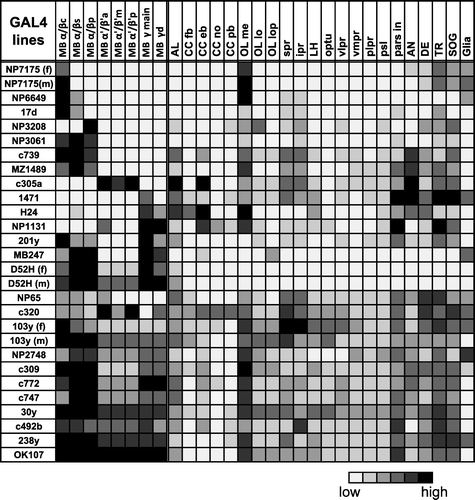
Figure 2 Drivers labeling the α/β neurons. Stereopairs show reconstructions of MB-GAL4s preferentially labeling the α/β neurons. The applied color illustrates the depth (see scale bar [25 µm] for the color code). Because of the space limitation, labels of the neuropils have been omitted from all figures. See also original confocal stacks for detail (http://mushroombody.net). Expression pattern was indistinguishable between males and females, unless stated. (A) Female NP7175 exclusively innervated the α/βc lobes in the MB. It also labeled the subesophageal ganglion, the cells in the medulla, and the processes in the tritocerebrum. See A for the expression pattern in the male. (B) NP6649 strongly labeled the α/βc, while the expression in the α/βs was weak. As in NP7175, the cells in the medulla were strongly labeled. It also has expression in the dorsal giant interneuron (DGI) (Ito et al., Citation1997). (C) As in NP6649, the α/βc were strongly labeled in 17d, while the expression in the α/βs was weak. In addition, weak labeling was occasionally detected in the pars intercerebralis, cells located around the subesophageal ganglion, DGI, and large neurons located around the calyx and protocerebral bridge. One very large neuron located ventrally to the calyx projects anteriorly and terminates in the superior and inferior protocerebrum. Collaterals of this neuron may also project to the deutocerebrum. Similar neurons were labeled by NP3208, c739, MZ1489, NP65, and c492b. Especially in MZ1489, their huge cell bodies are clearly visible (see G). (D) NP3208 specifically innervated the α/βp. The accessory calyx is visible as a protrusion originating from the dorsal calyx. The paired giant neurons located ventral to the calyx (see also the legend of C) and processes in the subesophageal ganglion were labeled. Further faint signals were detected in many neuropils, including the optic lobes. (E) NP3061 labeled all over the α/β lobes. Occasionally, barely detectable signals were in the pars intercerebralis, DGI, tritocerebrum, and subesophageal ganglion. NP3061 contained the least expression outside the MB in all the MB-GAL4s analyzed here. (F) c739 strongly innervated the entire α/β lobes. Outside the MB, it labeled elements in a wide range of neuropils, including local interneurons in the antennal lobe, the antennal nerve, ellipsoid body, a cluster of neurons projecting from/to the optic lobes, and many other cells throughout the brain (see also ). (G) MZ1489 innervated the entire α/β lobe, though the signal in the α/βc was less intense. Outside of the MB, it labeled a wide range of neuropils, including the pars intercerebralis, many cells in the medulla, antennal nerve, paired giant neurons located ventral to the calyx (see also the legend of C), and many other cells throughout the brain (see also ).
![Figure 2 Drivers labeling the α/β neurons. Stereopairs show reconstructions of MB-GAL4s preferentially labeling the α/β neurons. The applied color illustrates the depth (see scale bar [25 µm] for the color code). Because of the space limitation, labels of the neuropils have been omitted from all figures. See also original confocal stacks for detail (http://mushroombody.net). Expression pattern was indistinguishable between males and females, unless stated. (A) Female NP7175 exclusively innervated the α/βc lobes in the MB. It also labeled the subesophageal ganglion, the cells in the medulla, and the processes in the tritocerebrum. See Figure 7A for the expression pattern in the male. (B) NP6649 strongly labeled the α/βc, while the expression in the α/βs was weak. As in NP7175, the cells in the medulla were strongly labeled. It also has expression in the dorsal giant interneuron (DGI) (Ito et al., Citation1997). (C) As in NP6649, the α/βc were strongly labeled in 17d, while the expression in the α/βs was weak. In addition, weak labeling was occasionally detected in the pars intercerebralis, cells located around the subesophageal ganglion, DGI, and large neurons located around the calyx and protocerebral bridge. One very large neuron located ventrally to the calyx projects anteriorly and terminates in the superior and inferior protocerebrum. Collaterals of this neuron may also project to the deutocerebrum. Similar neurons were labeled by NP3208, c739, MZ1489, NP65, and c492b. Especially in MZ1489, their huge cell bodies are clearly visible (see G). (D) NP3208 specifically innervated the α/βp. The accessory calyx is visible as a protrusion originating from the dorsal calyx. The paired giant neurons located ventral to the calyx (see also the legend of C) and processes in the subesophageal ganglion were labeled. Further faint signals were detected in many neuropils, including the optic lobes. (E) NP3061 labeled all over the α/β lobes. Occasionally, barely detectable signals were in the pars intercerebralis, DGI, tritocerebrum, and subesophageal ganglion. NP3061 contained the least expression outside the MB in all the MB-GAL4s analyzed here. (F) c739 strongly innervated the entire α/β lobes. Outside the MB, it labeled elements in a wide range of neuropils, including local interneurons in the antennal lobe, the antennal nerve, ellipsoid body, a cluster of neurons projecting from/to the optic lobes, and many other cells throughout the brain (see also Figure 1). (G) MZ1489 innervated the entire α/β lobe, though the signal in the α/βc was less intense. Outside of the MB, it labeled a wide range of neuropils, including the pars intercerebralis, many cells in the medulla, antennal nerve, paired giant neurons located ventral to the calyx (see also the legend of C), and many other cells throughout the brain (see also Figure 1).](/cms/asset/d7d55611-706d-442a-9bef-20e421ac840d/ineg_a_347339_f0002_b.jpg)
Figure 3 Driver labeling the α′/β′ neurons. Stereopair shows a reconstruction of c305a preferentially labeling the entire α′/β′ lobes. The applied color illustrates the depth (see scale bar [25 µm] for the color code). See also original confocal stacks for detail (http://mushroombody.net). Outside the MB, it labeled broad neuropils, including the local interneurons in the antennal lobe, antennal nerve, ellipsoid body, large paired neurons originating from the subesophageal ganglion, and many other cells throughout the brain (see also ).
![Figure 3 Driver labeling the α′/β′ neurons. Stereopair shows a reconstruction of c305a preferentially labeling the entire α′/β′ lobes. The applied color illustrates the depth (see scale bar [25 µm] for the color code). See also original confocal stacks for detail (http://mushroombody.net). Outside the MB, it labeled broad neuropils, including the local interneurons in the antennal lobe, antennal nerve, ellipsoid body, large paired neurons originating from the subesophageal ganglion, and many other cells throughout the brain (see also Figure 1).](/cms/asset/9b3129d3-7112-47ba-9239-1e55ae2f3867/ineg_a_347339_f0003_b.jpg)
Figure 4 Drivers labeling the γ neurons. (A, B) Stereopairs show reconstructions of MB-GAL4s preferentially labeling the γ lobe. The applied color illustrates the depth (see scale bar [25 µm] for the color code). See also original confocal stacks for detail (http://mushroombody.net). (A) In the MB, expression of 1471 was restricted in the γ neurons. Expression in the γd neurons was very weak, if any. It labeled a broad range of neuropils outside of the MB, including the pars intercerebralis, antennal lobe, antennal nerve, and sensory nerves. Large paired neurons located ventrally to the SOG and projecting to the deutocerebrum and/or tritocerebrum were also labeled. Similar neurons are also found in NP65, H24, and 201y. (B) H24 strongly labeled the γ neurons, with extremely weak additional expression in the α/β neurons. Outside the MB, local interneurons in the antennal lobe, the medulla, ellipsoid body, deutocerebrum, and large paired neurons located ventrally to the subesophageal ganglion showed the reporter signals. (C–G) Frontal views of the projections of three consecutive confocal slices, including the γd lobe. In the left panels, the GAL4-positive processes (green) are shown with counterstaining of neuropils (anti-Synapsin; magenta). The right panels show only the reporter channel of the same stacks (black). In these sections, the γ lobe occupies the most dorsal part, while the β lobe occupies the most ventral part. The β′ lobe typically lies in between them. Arrows indicate the medial end of the γd subdivision. 1471 labeled the main part of the γ lobe, but not the γd subdivision (C), whereas the expression both in the γd subdivision and the main part of the γ lobe was seen in H24 and MB247 (D and E). (F) c320 labeled the γd subdivision without labeling the main γ lobe. (G) c305a specifically labeled the α′/β′ lobe, but not the γd subdivision. Scale bar=25 µm for C–G.
![Figure 4 Drivers labeling the γ neurons. (A, B) Stereopairs show reconstructions of MB-GAL4s preferentially labeling the γ lobe. The applied color illustrates the depth (see scale bar [25 µm] for the color code). See also original confocal stacks for detail (http://mushroombody.net). (A) In the MB, expression of 1471 was restricted in the γ neurons. Expression in the γd neurons was very weak, if any. It labeled a broad range of neuropils outside of the MB, including the pars intercerebralis, antennal lobe, antennal nerve, and sensory nerves. Large paired neurons located ventrally to the SOG and projecting to the deutocerebrum and/or tritocerebrum were also labeled. Similar neurons are also found in NP65, H24, and 201y. (B) H24 strongly labeled the γ neurons, with extremely weak additional expression in the α/β neurons. Outside the MB, local interneurons in the antennal lobe, the medulla, ellipsoid body, deutocerebrum, and large paired neurons located ventrally to the subesophageal ganglion showed the reporter signals. (C–G) Frontal views of the projections of three consecutive confocal slices, including the γd lobe. In the left panels, the GAL4-positive processes (green) are shown with counterstaining of neuropils (anti-Synapsin; magenta). The right panels show only the reporter channel of the same stacks (black). In these sections, the γ lobe occupies the most dorsal part, while the β lobe occupies the most ventral part. The β′ lobe typically lies in between them. Arrows indicate the medial end of the γd subdivision. 1471 labeled the main part of the γ lobe, but not the γd subdivision (C), whereas the expression both in the γd subdivision and the main part of the γ lobe was seen in H24 and MB247 (D and E). (F) c320 labeled the γd subdivision without labeling the main γ lobe. (G) c305a specifically labeled the α′/β′ lobe, but not the γd subdivision. Scale bar=25 µm for C–G.](/cms/asset/b9906eb9-dc17-4143-ad90-c2430c8bf352/ineg_a_347339_f0004_b.jpg)
Figure 5 Drivers labeling multiple types of Kenyon cells. Stereopairs show reconstructions of MB-GAL4s preferentially labeling the multiple types of Kenyon cells. The applied color illustrates the depth (see scale bar [25 µm] for the color code). Some lines exhibited detectable expression in all types of Kenyon cells, but were categorized into this group, because expression in certain subdivisions was remarkably weak. See also original confocal stacks for detail (http://mushroombody.net). (A) NP1131 labeled the α′/β′a and γ lobes. Additional expression was seen in the ellipsoid body, subesophageal ganglion, pars intercerebralis, large interneurons connecting the optic lobe and the central brain, and other cells distributed in the brain (see also ). (B) 201y labeled the α/βc and γ neurons. Outside the MB, additional expression was seen in the several glomeruli in the antenna lobe, pars intercerebralis, large paired neurons located ventral to the subesophageal ganglion, DGI, and other neurons. (C) MB247 strongly labeled the α/β and γ neurons with very low background expression. Expression in the α/βc was weaker than in the other α/β subdivisions. Additional expression was detected in the cells in the lobula plate and surface glia. (D) Female D52H strongly labeled the α/β and γ neurons with very low background expression. Expression in the α/βc was weaker than in the other α/β subdivisions. See B for the expression pattern in the male. (E) NP65 labeled the α/βs, α/βc, and α′/β′a neurons. We observed reporter signals also in the antennal lobe, deutocerebrum, tritocerebrum, DGI, large paired neurons located ventral to the calyx (see also the legend of C), two large neurons projecting medially to the subesophageal ganglion, and many cells throughout the central brain. (F) In c320, the α/β, α′/β′, and γd lobes were labeled (F). Expression in the α′/β′m neurons was weaker than the rest of α′/β′ subdivisions. Outside the MB, it labeled the majority of neuropils. Among them, relatively strong expressions were observed in surface glia, the protocerebral bridge, subesophageal ganglion, optic lobes, and superior protocerebrum (see also ). (G) Female 103y labeled the α/β neurons strongly. Outside the MB, it labeled the optic lobes, inferior and superior protocerebrum, cells on the subesophageal ganglion, surface glia, and many cells in the posterior cortex. See C for expression pattern in the male. (H) In NP2748, weak reporter signals were observed in all of the lobes. It also labeled the medulla, pars intercerebralis, DGI, surface glia, and many other cells.
![Figure 5 Drivers labeling multiple types of Kenyon cells. Stereopairs show reconstructions of MB-GAL4s preferentially labeling the multiple types of Kenyon cells. The applied color illustrates the depth (see scale bar [25 µm] for the color code). Some lines exhibited detectable expression in all types of Kenyon cells, but were categorized into this group, because expression in certain subdivisions was remarkably weak. See also original confocal stacks for detail (http://mushroombody.net). (A) NP1131 labeled the α′/β′a and γ lobes. Additional expression was seen in the ellipsoid body, subesophageal ganglion, pars intercerebralis, large interneurons connecting the optic lobe and the central brain, and other cells distributed in the brain (see also Figure 1). (B) 201y labeled the α/βc and γ neurons. Outside the MB, additional expression was seen in the several glomeruli in the antenna lobe, pars intercerebralis, large paired neurons located ventral to the subesophageal ganglion, DGI, and other neurons. (C) MB247 strongly labeled the α/β and γ neurons with very low background expression. Expression in the α/βc was weaker than in the other α/β subdivisions. Additional expression was detected in the cells in the lobula plate and surface glia. (D) Female D52H strongly labeled the α/β and γ neurons with very low background expression. Expression in the α/βc was weaker than in the other α/β subdivisions. See Figure 7B for the expression pattern in the male. (E) NP65 labeled the α/βs, α/βc, and α′/β′a neurons. We observed reporter signals also in the antennal lobe, deutocerebrum, tritocerebrum, DGI, large paired neurons located ventral to the calyx (see also the legend of Figure 2C), two large neurons projecting medially to the subesophageal ganglion, and many cells throughout the central brain. (F) In c320, the α/β, α′/β′, and γd lobes were labeled (Figure 3F). Expression in the α′/β′m neurons was weaker than the rest of α′/β′ subdivisions. Outside the MB, it labeled the majority of neuropils. Among them, relatively strong expressions were observed in surface glia, the protocerebral bridge, subesophageal ganglion, optic lobes, and superior protocerebrum (see also Figure 1). (G) Female 103y labeled the α/β neurons strongly. Outside the MB, it labeled the optic lobes, inferior and superior protocerebrum, cells on the subesophageal ganglion, surface glia, and many cells in the posterior cortex. See Figure 7C for expression pattern in the male. (H) In NP2748, weak reporter signals were observed in all of the lobes. It also labeled the medulla, pars intercerebralis, DGI, surface glia, and many other cells.](/cms/asset/b7381959-8a07-4e99-802c-4115fa48c907/ineg_a_347339_f0005_b.jpg)
Figure 6 GAL4 strains labeling all of the lobes. Stereopairs show reconstructions of MB-GAL4s labeling all of the lobes. The applied color illustrates the depth (see scale bar [25 µm] for the color code). Since most of the lines in this category exhibited expression in many neuropils, we describe only strong signals here (see also ). See also original confocal stacks for detail (http://mushroombody.net). (A) In the MB, c309 strongly labeled the α/β and γ neurons. We also observed very weak expression in the α′/β′ neurons. Outside the MB, the majority of the labeled neuropils included the antennal lobe, tritocerebrum, subesophageal ganglion, and optic lobes (see also ). Additionally, many different sensory nerves and the cervical connectives were stained. (B) c772 preferentially labeled the α/βp, α/βs, and γ neurons with weaker expression in the α/βc and α′/β′ neurons. It also labeled the optic lobes, ventrolateral protocerebrum, local interneurons in the antennal lobe, and many cells on the subesophageal ganglion. (C) As in c772, c747 labeled the α/βp, α/βs, and γ neurons strongly and the α/βc and α′/β′ neurons weakly. Outside the MB, we observed reporter signals in the optic lobes, antennal nerve, local interneurons in the antennal lobe, pars intercerebralis, and many cells on the subesophageal ganglion. (D) In 30y, all the MB subdivisions were innervated. It also labeled the optic lobes, antennal lobe, pars intercerebralis, and many cells surrounding the subesophageal ganglion. It also labeled a cluster of small neurons that are located dorsolateral to the optic tubercle and that project first ventrally and extend laterally toward the dorsolateral edge of the ventrolateral protocerebrum. Similar neurons were observed in 238y. (E) c492b labeled all types of Kenyon cells, although expression in the γd neurons was relatively weak. Compared to other lines in this category, expressions outside the MB were less pronounced. We observed reporter signals in the pars intercerebralis, local inter neurons in the antennal lobe, deutocerebrum, subesophageal ganglion, and large paired neurons located ventral to the calyx (see also the legend of C). (F) All types of the Kenyon cells were strongly labeled by 238y. Outside the MB, this line labeled the optic lobe, superior protocerebrum, pars intercerebralis, and many neurons surrounding the entire subesophageal ganglion. (G) In OK107, all the subdivisions of the MB were strongly and uniformly labeled. Outside the MB, we observed strong reporter signals in the optic lobe, antennal lobe, pars intercerebralis, and cells on the subesophageal ganglion.
![Figure 6 GAL4 strains labeling all of the lobes. Stereopairs show reconstructions of MB-GAL4s labeling all of the lobes. The applied color illustrates the depth (see scale bar [25 µm] for the color code). Since most of the lines in this category exhibited expression in many neuropils, we describe only strong signals here (see also Figure 1). See also original confocal stacks for detail (http://mushroombody.net). (A) In the MB, c309 strongly labeled the α/β and γ neurons. We also observed very weak expression in the α′/β′ neurons. Outside the MB, the majority of the labeled neuropils included the antennal lobe, tritocerebrum, subesophageal ganglion, and optic lobes (see also Figure 1). Additionally, many different sensory nerves and the cervical connectives were stained. (B) c772 preferentially labeled the α/βp, α/βs, and γ neurons with weaker expression in the α/βc and α′/β′ neurons. It also labeled the optic lobes, ventrolateral protocerebrum, local interneurons in the antennal lobe, and many cells on the subesophageal ganglion. (C) As in c772, c747 labeled the α/βp, α/βs, and γ neurons strongly and the α/βc and α′/β′ neurons weakly. Outside the MB, we observed reporter signals in the optic lobes, antennal nerve, local interneurons in the antennal lobe, pars intercerebralis, and many cells on the subesophageal ganglion. (D) In 30y, all the MB subdivisions were innervated. It also labeled the optic lobes, antennal lobe, pars intercerebralis, and many cells surrounding the subesophageal ganglion. It also labeled a cluster of small neurons that are located dorsolateral to the optic tubercle and that project first ventrally and extend laterally toward the dorsolateral edge of the ventrolateral protocerebrum. Similar neurons were observed in 238y. (E) c492b labeled all types of Kenyon cells, although expression in the γd neurons was relatively weak. Compared to other lines in this category, expressions outside the MB were less pronounced. We observed reporter signals in the pars intercerebralis, local inter neurons in the antennal lobe, deutocerebrum, subesophageal ganglion, and large paired neurons located ventral to the calyx (see also the legend of Figure 2C). (F) All types of the Kenyon cells were strongly labeled by 238y. Outside the MB, this line labeled the optic lobe, superior protocerebrum, pars intercerebralis, and many neurons surrounding the entire subesophageal ganglion. (G) In OK107, all the subdivisions of the MB were strongly and uniformly labeled. Outside the MB, we observed strong reporter signals in the optic lobe, antennal lobe, pars intercerebralis, and cells on the subesophageal ganglion.](/cms/asset/5e34e8bb-2796-4b14-bcc8-6d369daf65b3/ineg_a_347339_f0006_b.jpg)
Figure 7 GAL4 strains with sex-dependent difference. Stereopairs show reconstructions of MB-GAL4s with sex-specific reporter expression. The applied color illustrates the depth (see scale bar [25 µm] for the color code). See also original confocal stacks for detail (http://mushroombody.net). (A) Compared to the females, male NP7175 labeled slightly broader α/βc ( A). In addition to the background expression seen in the female, surface glia was strongly labeled. (B) In contrast to the female, male D52H additionally labeled the α′/β′ neurons. Moreover, the innervation of one glomerulus by the olfactory receptor neurons was more pronounced in the male. Otherwise, background expression was unusually low as in females. (C) Male 103y labeled all subtypes of Kenyon cells, whereas in the female, the expression in the α′/β′ and γ lobes was very faint (see G). In addition, expression in surface glia and dense terminals in the superior medial protocerebrum were less pronounced in males. Outside the MB, it labeled the processes in the medulla and lobula, middle superior lateral protocerebrum, local interneurons in the antennal lobe, and many neurons supplying the subesophageal ganglion and tritocerebrum.
![Figure 7 GAL4 strains with sex-dependent difference. Stereopairs show reconstructions of MB-GAL4s with sex-specific reporter expression. The applied color illustrates the depth (see scale bar [25 µm] for the color code). See also original confocal stacks for detail (http://mushroombody.net). (A) Compared to the females, male NP7175 labeled slightly broader α/βc (Figure 2 A). In addition to the background expression seen in the female, surface glia was strongly labeled. (B) In contrast to the female, male D52H additionally labeled the α′/β′ neurons. Moreover, the innervation of one glomerulus by the olfactory receptor neurons was more pronounced in the male. Otherwise, background expression was unusually low as in females. (C) Male 103y labeled all subtypes of Kenyon cells, whereas in the female, the expression in the α′/β′ and γ lobes was very faint (see Figure 5G). In addition, expression in surface glia and dense terminals in the superior medial protocerebrum were less pronounced in males. Outside the MB, it labeled the processes in the medulla and lobula, middle superior lateral protocerebrum, local interneurons in the antennal lobe, and many neurons supplying the subesophageal ganglion and tritocerebrum.](/cms/asset/e3f60927-f8be-45e2-82a0-e3c066b8955f/ineg_a_347339_f0007_b.jpg)
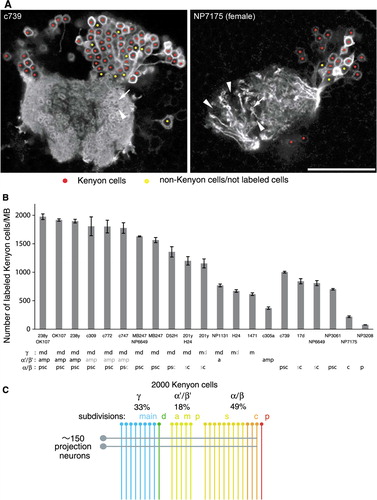
Figure 8 The number of genetically labeled Kenyon cells. (A) Single confocal sections through the calyx and cell-body cluster of Kenyon cells as representative pictures of the counting procedure (c739 and NP7175). Nuclei of Kenyon cells are marked with red, while non-Kenyon cells, unlabeled cells, or spaces between cells are marked with yellow. The arrow and arrowhead indicate microglomeruli innervated and not innervated by labeled Kenyon cells, respectively. Scale bar=25 µm. (B) The numbers of genetically labeled Kenyon cells in different MB-GAL4 drivers of the females. Labeled Kenyon cell subtypes are indicated below (see also ). The label in gray shows weak expression. See the legend of for abbreviations. N=5–8. Error bars represent standard error of the mean. (C) Model of the numerical composition of the Drosophila MB. The γ, α′/β′, and α/β neurons, respectively, contribute to 33, 18, and 49% of∼2,000 Kenyon cells. One circle and line represents∼75 cells. Approximately 150 iACT projections neurons terminate on the Kenyon cells, except for the α/βp neurons (rightmost line).