Figures & data
Figure 1. (a) GABA and glutamate-gated chloride channels share similar structures with individual subunits comprising a large extracellular N-terminal domain (for ligand binding) and four transmembrane domains (M1–M4). The mature receptor (b) is a pentamer and gates a chloride-selective channel following GABA/Glu binding. The positions of mutations implicated in cyclodiene/fipronil resistance at the GABA-R (red circles) and avermectin resistance at the GluCl (yellow circles) are shown in (a) and the corresponding regions of the mature receptors highlighted in (b). Structural models of these binding sites (AVE, avermectin and FIP, fipronil) are shown in (c) (adapted from Casida and Durkin (Casida & Durkin, Citation2015).
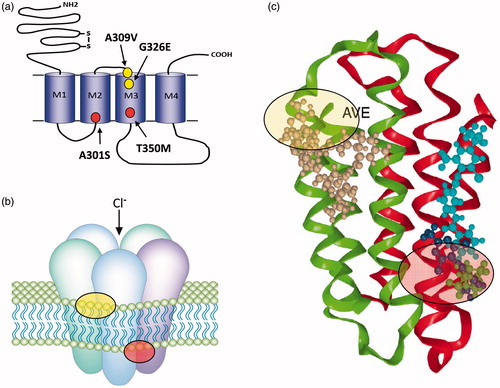
Figure 2. (a) nAChRs share a similar overall structure to the ligand-gated chloride channels with individual subunits containing four trans-membrane domains (M1–M4) and a large extracellular N-terminal domain. The mature receptor (b) is a pentamer of either identical or non-identical subunits and gates a cation-selective channel following acetylcholine binding to the extracellular domain. Neonicotinoids such as imidacloprid activate the channel by binding to the same site. The only field-confirmed case of target site resistance to these compounds involves mutation (red circle) of arginine 81 to threonine (R81T) in Myzus persicae β1 subunit that causes repulsion of imidacloprid binding at the α/β subunit interface as shown in (c) (taken from Bass et al. (Citation2015)).
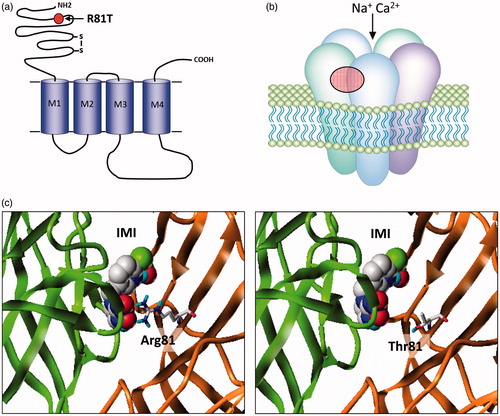
Figure 3. The voltage-gated sodium channel as a target for pyrethroids. (a) Diagram of the voltage gated sodium channel showing the four repeat domains (I-IV), each comprising six membrane spanning helices (S1-S6). The positions of resistance mutations that identify pyrethroid binding sites PyR1 (red circles) and PyR2 (blue circles) are highlighted. The proposed PyR1 (b) and PyR2 (c) sites are shown in more detail, taken from O'Reilly et al. (Citation2006) and Du et al. (Citation2013). Positions of the mutations originally identified in pyrethroid and DDT-resistant parats Drosophila strains are also shown (yellow circles).
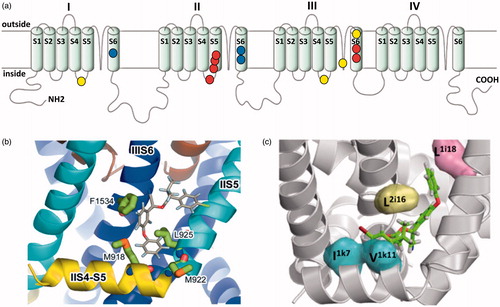
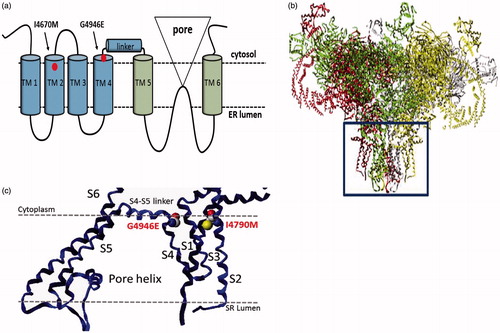
Figure 4. Structure of the insect ryanodine receptor and the location of resistance associated mutations. (a) Topology of the C-terminal domain of the insect RyR showing the six membrane spanning helices (TM1–6) with the positions of the two resistance mutations (I4790M & G4946E) that map to this region of the channel highlighted. (b) Shows a full model of the insect RyR based on the recently reported structure of a rabbit RyR, with the membrane domain (boxed) further magnified in (c) to show the close proximity of the resistance-associated mutations (adapted from (Steinbach et al., Citation2015)).