Figures & data
Figure 1. Crossing scheme for conducting genetic swaps. An example of the crossing strategy to genetically swap the original InSITE Gal4 inserted on the third chromosome with a donor inserted on the second chromosome.
Figure 2. GAL4 expression patterns of InSITE lines subjected to genetic swap. Projections of confocal stacks. Adult brains were stained for mCD8 (green) and the neuropil marker, Bruchpilot (Brp, magenta). D, dorsal; L, lateral. Scale bar, 20 μm.
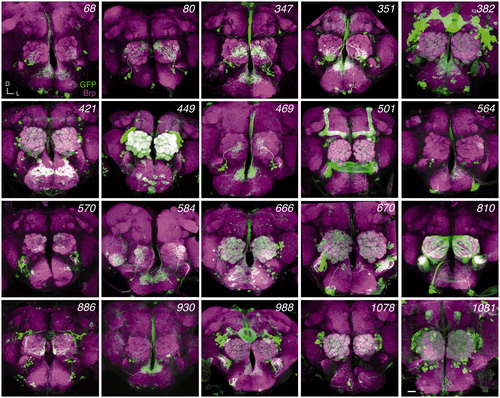
Table 1. Summary of genetic swap results.
Figure 3. Genetic swap of GAL4 enhancer trap to LexA. (a) Primer sets used to verify the genetic conversion of GAL4 to LexA. (b) PCR analysis to confirm the swap results. Genomic DNA samples: lane 1, 584-GAL4; lane 2, 670-GAL4; lane 3, LexAL19.2L donor; lane 4, LexAL44.2 donor; lane 5, water; lane 6, 584-LexAL19.2L (2-4); lane 7, 584-LexAL44.2 (1-1); lane 8, 670-LexAL44.2 (28-3B8); lane 9, 670-LexAL44.2 (28-3C3); lane 10, 670-LexAL44.2 (28-3D3). The asterisk denotes non-specific PCR product. (c–e) Functional validation of swapped 564-LexA (c,d) and 670-LexA (e) lines. Adult brains were stained for mCD8 or GFP (green) and Bruchpilot (Brp, magenta). (d) Low magnification images of (c) are shown. Arrows indicate optic lobes. Image of 670-GAL4 is reproduced from . Scale bars, 20 μm (c, e) or 50 μm (d).
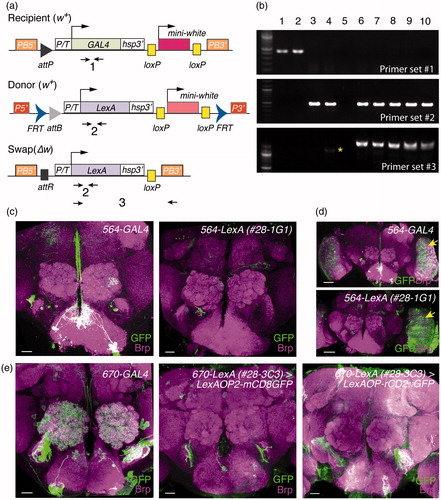
Figure 4. Genetic swap of GAL4 enhancer trap to GAL80. (a) Primer sets used to verify the genetic conversion of GAL4 to GAL80. (b) PCR analysis to confirm the swap results. Genomic DNA samples: lane 1, 347-GAL4; lane 2, 449-GAL4; lane 3, GAL80E22.1 donor; lane 4, water; lane 5, 347-GAL80 (1-3); lane 6, 347-GAL80 (4-4); lane 7, 449-GAL80 (2-1); lane 8, 449-GAL80 (3-1). (c–e) Functional validation of swapped GAL80 lines. Adult brains were stained for mCD8 (green) and Bruchpilot (Brp, magenta). (c) Compared to the processes of GFP-positive cells in the 347-GAL4 AL (yellow arrowheads, left panel), 347-GAL80 appears to have fewer GFP-positive processes marking 347-positive neurons in the AL, with additional somas and innervations observed (cyan arrowheads, right panel). Although 449-GAL80 did not obviously suppress 449-GAL4 expressions (d), it did suppress GAL4 activity in a subset of krasavietz-positive LNs (arrowheads) (e). Scale bars, 20 μm. (f) Quantification of GFP-positive LNs or viPNs in different GAL4-GAL80 combinations. Bar graphs show mean ± S.D. Numbers of analyzed ALs are indicated in the parenthesis. Student’s t-test was used to compare the groups. *p<.05, **p<.01, ***p<.001.
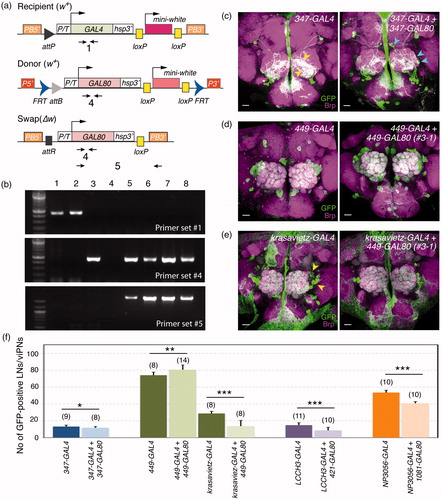
Figure 5. Validation of GAL4 to QF enhancer trap swaps. (a) Primer sets used to verify the genetic conversion of GAL4 to QF. (b) PCR analysis to confirm the swap results. Genomic DNA samples: lane 1, 449-GAL4; lane 2, 584-GAL4; lane 3, QF.Q10B donor; lane 4, water; lane 5, 449-QF.Q10B (11-1); lane 6, 449-QF.Q10B (14-2); lane 7, 584-QF.Q10B (1-1). Asterisks denote non-specific PCR products. (c–d) Functional validation of swapped QF lines. Adult brains were stained for mCD8 (green, left panels) or mtdT-3xHA (green, right panels) and Bruchpilot (Brp, magenta). Images of 449-GAL4 and 584-GAL4 are reproduced from . Scale bars, 20 μm.
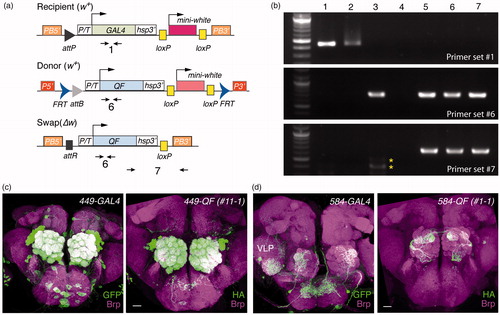
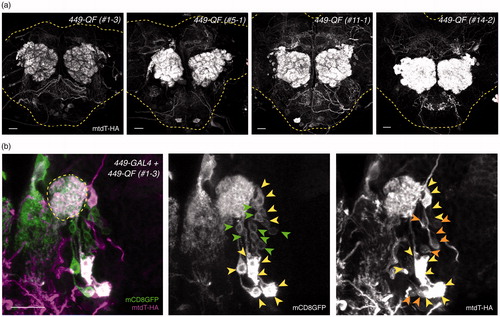
Figure 6. Differential expression levels in independent QF swap lines. (a) The QF expression patterns of four representative 449-QF lines, showing weak LN-QF, medium LN-QF and strong LN-QF expression. All brains were imaged with the same confocal configurations for mtdT-3xHA. Yellow dashed lines show brain contours based on Brp staining (not shown). (b) 449-GAL4-driven mCD8GFP (green) and 449-QF-driven mtdT-3xHA (magenta) were co-stained in a late third instar larval brain. LNs labeled only by 449-GAL4, only by 449-QF and by both 449-GAL4 and 449-QF are indicated by green, orange and yellow arrowheads, respectively. Scale bars, 20 μm.
Suplementary_information-rev1.pdf
Download PDF (3.3 MB)Data availability
The authors declare that all data supporting the findings of this study are available within the article and its supplementary information files, or from the corresponding author upon reasonable request.
