Figures & data
Figure 2. Comparison of high-resolution structural information of DEAD-box helicases. (a) Surface representation of DDX3 with AMP (PDB file: 5E7J); (b) Ribbon representation of helicase core domains of DDX3(grey) and DDX5(green) overlaid with their associated nucleotide phosphate, AMP and ADP, respectively (PDB files: 5E7J and 3FE2); (c) Coordinating residues with AMP within the Q and Walker I motifs (for DDX3). Surface (d) and ribbon (e) representation of helicase core domain structure of DDX3 presenting RNA-binding site (produced with comparative modelling of the helicase eI4A, PDB file: 2HYI). (f) DDX1 SPRY domain tertiary structure indicating two stacked concave β-sheets (pink and blue) with a third lower β-sheet (yellow) (PDB ID 4XW3). Images were produced using PyMOL (PyMOL https://pymol.org/2/).
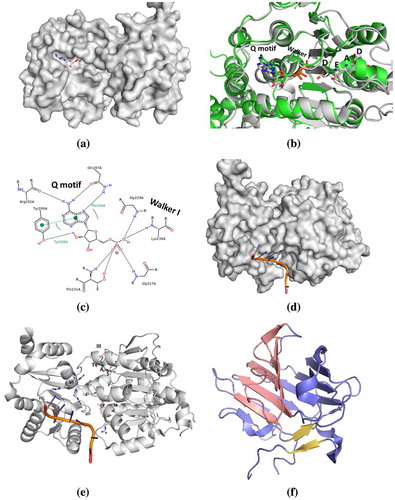
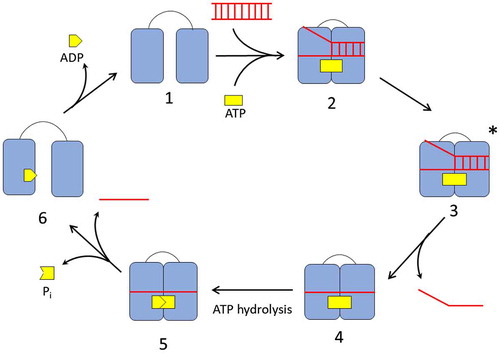
Figure 3. The catalytic cycle of DEAD-box helicase. In the absence of ligands, the DEAD-box helicase exists in an open conformation (1). Binding of RNA and ATP cause the helicase to switch to a closed conformation (2) aligning the ATP active site for hydrolysis and locally destabilizing the RNA in its binding site. In the activated complex (3), the first RNA strand dissociates. ATP hydrolysis (4) and release of the phosphate and second RNA strand (5) enables the helicase to re-open (6) and reset for the next cycle.