Figures & data
Table I. Nucleotide sequences of primers for target genes.
Figure 1. PF induced Hsp70 in a dose- and time-dependent manner. (a) SDS-PAGE and Western blotting were performed on U937 cells treated with PF for 30 min at different concentrations (80–640 μg/ml for dose-dependent experiments and 160 µg/ml for time-dependent experiments). After which cells were incubated at different times (0–12 h for time-dependent experiments and 3 h for dose-dependent experiment). The signals were visualized by a luminescent image analyser using an ECL system. (b,c) Bands were quantified by densitometry and normalized with β-actin. Data are presented as mean ± SD (n = 3). *p < 0.05 vs. control (Student's t test). Cont, control.
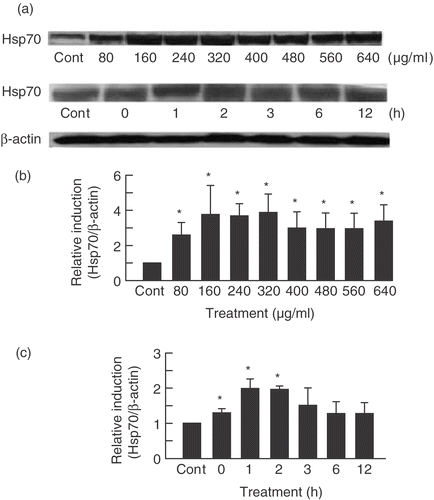
Table II. Upregulated genes under PF treatment.
Table III. Downregulated genes under PF treatment.
Figure 2. Genetic networks of genes associated with PF treatment. The cells were treated with PF (160 µg/ml, 30 min) and harvested after 3 h incubation. The genes associated with PF were analysed by the Ingenuity Pathway Analysis tool. The network is displayed graphically as nodes (genes) and edges (biological relationships between nodes).
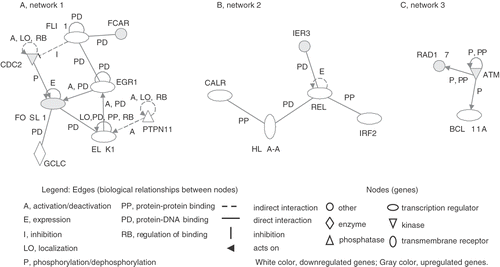
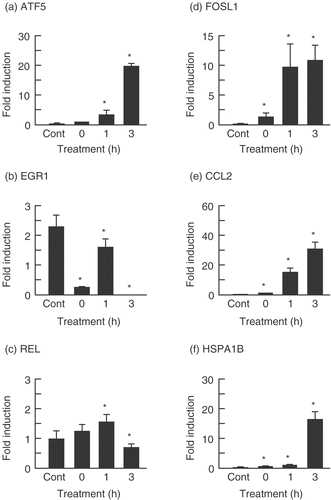
Figure 3. Verification of microarray results with real-time quantitative PCR assay. The cells were treated with PF (160 µg/ml, 30 min) and harvested after incubation for 0, 1, and 3 h at 37°C. Each mRNA expression level was normalized with GAPDH. Data are presented as mean ± SD (n = 4). (a) ATF5 (Activating transcription factor (5); (b) EGR1 (Early growth response 1); (c) REL (Reticuloendotheliosis viral oncogene homolog); (d) FOSL1 (FOS-like antigen 1); (e) CCL2 (Chemokine (C–C motif) ligand 2); (f) HSPA1 (Heat shock 70 kDa protein B). *p < 0.05 versus control (Student's t test). Cont, control.