Figures & data
Figure 1. Histograms of inferred from Langevin simulations are shown for
for
The gray bars represent the normalized counts
of
and the solid black line is the Gumbel distribution function (EquationEquation (9)
(9)
(9) ) with the corresponding value of location parameter
fitted for each case.
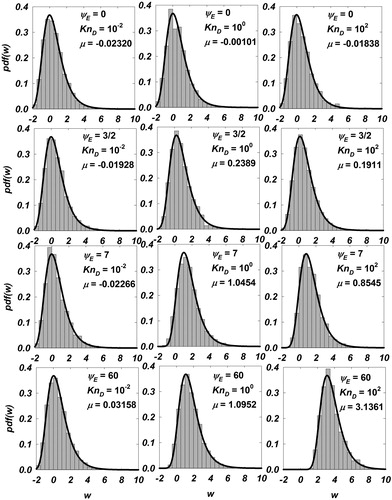
Figure 2. Fitted values of the location parameter for
for
shown as data points. The corresponding fit for
(EquationEquation (11)
(11)
(11) ) is shown as dashed lines of the same color.
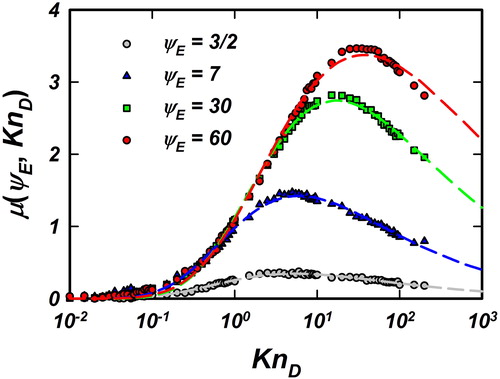
Figure 3. (a) Langevin-inferred (referred to as
) and prediction of EquationEquation (10)
(10)
(10) (dashed lines with the same color as the symbols) are plotted as a function of
for
Also shown are the infinitely collisional (EquationEquation (6a)
(6a)
(6a) ) and collisionless (EquationEquation (6b)
(6b)
(6b) ) limits as black dashed lines. (b) Plot showing the % difference between the prediction of EquationEquation (10)
(10)
(10) and
defined as
The same symbols are used in both plots to denote different values of
Reference lines at
are included to guide the eye. A compilation of more the datasets and their deviations from predictions of EquationEquation (10)
(10)
(10) are provided in Figure S3, SI.
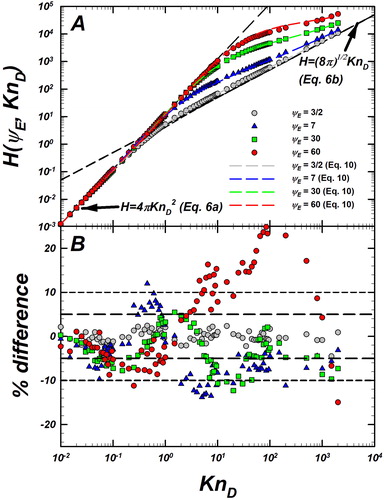
Figure 4. Plot showing the Langevin-inferred (referred to as
) for
Panels (a–c) present
for
as data points. The continuum limit (EquationEquation (6a)
(6a)
(6a) ) and free molecular limit (EquationEquation (6b)
(6b)
(6b) ) are shown as black dotted lines. Also shown is the hard sphere fit (EquationEquation (8)
(8)
(8) ) and the unscreened Coulomb model (EquationEquation (10)
(10)
(10) ). The common legend for panels (a–c) is given in panel (b). Panels (d–f) plot the comparison of the difference between the Langevin-inferred
(referred to as
) and (1) the prediction of the hard sphere fit (referred to as
) for
shown as blue triangles and (2) the prediction of Coulomb model (referred to as
) for
shown as green circles. Reference lines for denoting difference levels of
and
are shown to guide the eye. The common legend for panels (d–f) is given at the bottom.
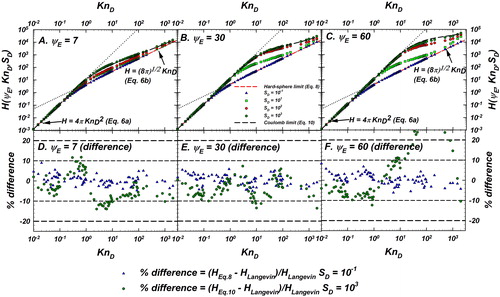
Figure 5. Langevin-inferred (referred to as
shown as green circles with black outline) and the predictions of various models considered for evaluation are plotted as a function of
for
In each panel, the predictions of the unscreened Coulomb (
) model (EquationEquation (10)
(10)
(10) , blue dashed line), D’Yachkov et al. (Citation2007) in orange, Gatti and Kortshagen (Citation2008) in pink, Gopalakrishnan and Hogan (Citation2012) in dark green, and Zobnin et al. (Citation2008) in red are shown. Also shown are the infinitely collisional (EquationEquation (6a)
(6a)
(6a) ) and collision-less (EquationEquation (6b)
(6b)
(6b) ) limits as dashed black lines. For comparison of the difference between model predictions and
see .
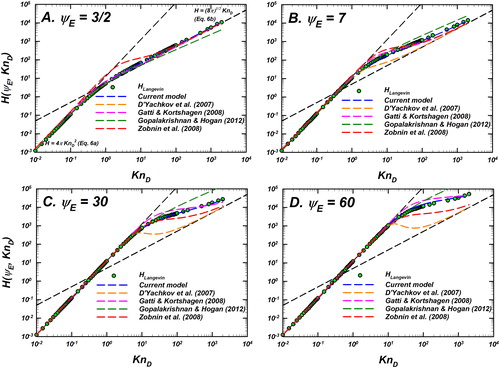
Figure 6. Comparison of the difference between the Langevin-inferred (referred to as
) and the prediction of various models as noted in the legend for
as a function of
Differences of the models considered are shown as dashed lines including the current model (EquationEquation (10)
(10)
(10) with
) in blue, D’Yachkov et al. (Citation2007) in orange, Gatti and Kortshagen (Citation2008) in pink, Gopalakrishnan and Hogan (Citation2012) in dark green, and Zobnin et al. (Citation2008) in red. The y-axis shows the % difference between the predictions of each model and
% difference is defined as
Reference lines for denoting difference levels of
and
are shown to guide the eye. This plot is to be read in conjunction with .
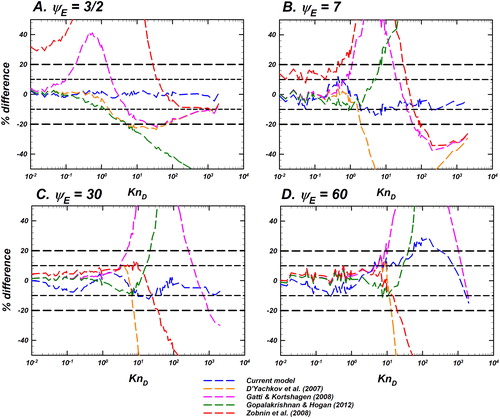
Figure 7. Langevin-inferred (referred to as
shown as green circles with black outline) and the predictions of various models considered for evaluation are plotted as a function of
for
and a fixed
In each panel, the predictions of the unscreened Coulomb model (EquationEquation (10)
(10)
(10) , blue dashed line), Gatti and Kortshagen (Citation2008) in pink, Zobnin et al. (Citation2008) in red and Hutchinson and Patacchini (Citation2007) in gray are shown. Also shown are the infinitely collisional (EquationEquation (6a)
(6a)
(6a) ) and collision-less (EquationEquation (6b)
(6b)
(6b) ) limits as black dashed lines. For comparison of the difference between model predictions and
see .
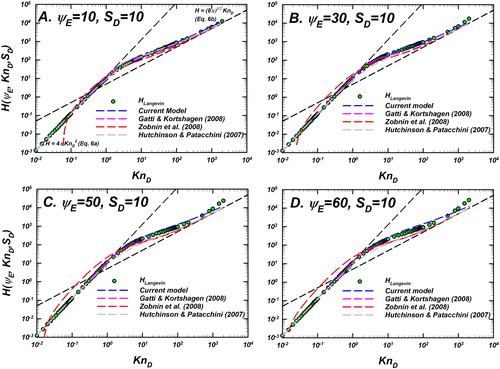
Figure 8. Comparison of the difference between the Langevin-inferred (referred to as
) and the prediction of various models as noted in the legend for
and a fixed
Differences of the models considered are shown as dashed lines including the current model (EquationEquation (10)
(10)
(10) ) in blue, Gatti and Kortshagen (Citation2008) in pink, Zobnin et al. (Citation2008) in red, and Hutchinson and Patacchini (Citation2007) in gray. The y-axis shows the % difference between the predictions of each model and
% difference is defined as
Reference lines for denoting difference levels of
and
are shown to guide the eye. This plot is to be read in conjunction with .
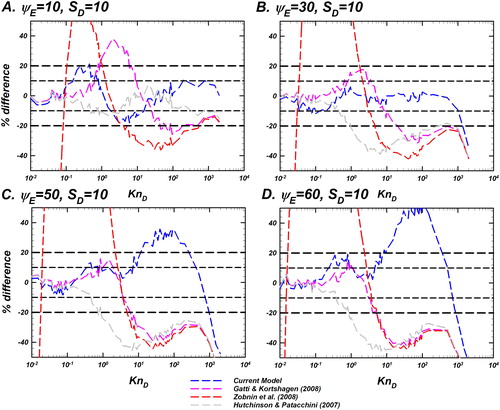
Figure 9. Langevin-inferred (referred to as
shown as green circles with black outline) and the predictions of various models considered for evaluation are plotted as a function of
for
and a fixed
In each panel, the predictions of the unscreened Coulomb model (EquationEquation (10)
(10)
(10) , blue dashed line), Gatti and Kortshagen (Citation2008) in pink, Zobnin et al. (Citation2008) in red, and Hutchinson and Patacchini (Citation2007) in gray are shown. Also shown are the infinitely collisional (EquationEquation (6a)
(6a)
(6a) ) and collision-less (EquationEquation (6b)
(6b)
(6b) ) limits as black dashed lines. For comparison of the difference between model predictions and
see .
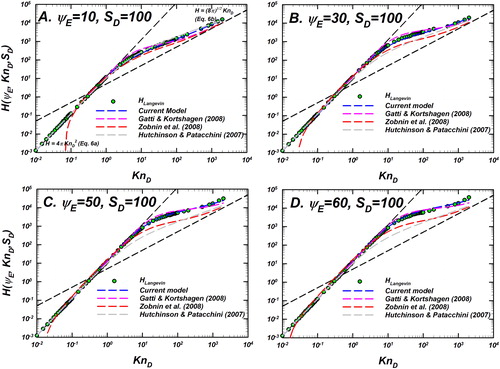
Figure 10. Comparison of the difference between the Langevin-inferred (referred to as
) and the prediction of various models as noted in the legend for
and a fixed
Differences of the models considered are shown as dashed lines including the current model (EquationEquation (10)
(10)
(10) ) in blue, Gatti and Kortshagen (Citation2008) in pink, Zobnin et al. (Citation2008) in red, and Hutchinson and Patacchini (Citation2007) in gray. The y-axis shows the % difference between the predictions of each model and
% difference is defined as
Reference lines for denoting difference levels of
and
are shown to guide the eye. This plot is to be read in conjunction with .
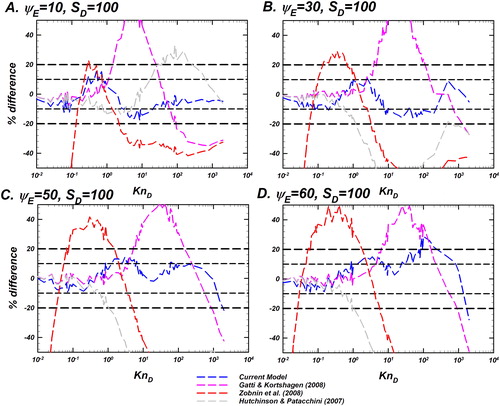
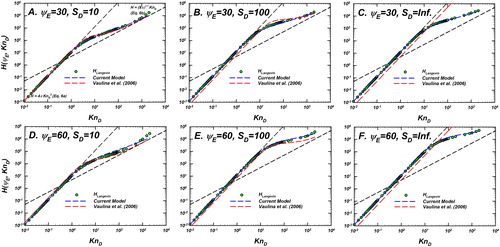
Figure 11. Langevin-inferred (referred to as
shown as green circles with black outline) and the predictions of Langevin dynamics based models—the current model and the model of Vaulina, Repin, and Petrov (Citation2006) are plotted as a function of
for
and
In each panel, the predictions of the current model (EquationEquation (10)
(10)
(10) , blue dashed line) and Vaulina, Repin, and Petrov (Citation2006) (red dashed line) are shown. Also shown are the infinitely collisional (EquationEquation (6a)
(6a)
(6a) ) and collision-less (EquationEquation (6b)
(6b)
(6b) ) limits as black dashed lines.