Figures & data
Figure 2. Multiplex PCR for detection of t(11;14) and t(14;18). M: molecular marker. Lane 1: cell line DOHH-2, lane 2: cell line WSU-NHL, lane 3: lymph node biopsy from patient no. 261, lane 4: cell line JVM-2, lane 5: blood from patient no. 28, and lane 6: ddH2O. Multiplex PCR: primers amplifying MBR1, MBR2, and MCR in t(14;18) as well as MTC and mTC1p94 in t(11;14). Split reactions: 5 primer sets detecting t(14;18) MBR1, t(14;18) MBR2, t(14;18) MCR, t(11;14) MTC, and t(11;14) mTC1p94. A control gene (TCF20) is co-amplified in all lanes. White arrows show the translocation positive PCR products in the split reactions, large black arrow exemplifies a PCR product of the control gene, while the thin black arrow exemplifies a weak germline PCR band.
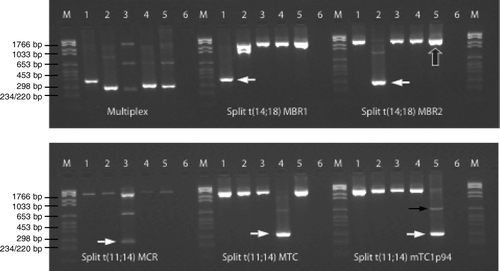
Table I. PCR positive for t(11;14). Comparison between PCR, flow cytometry, and histology at the same time of sampling.
Table II. PCR positive for t(14;18). Comparison between PCR, flow cytometry, and histology at the same time of sampling.
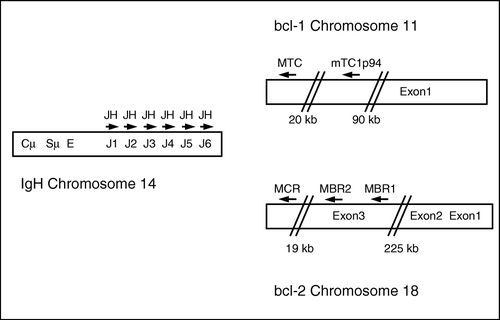
Figure 3. Breakpoints from the fusion DNA found in patients included in this study. Arrows indicate breakpoints and numbers refer to patient IDs. A. Sequence of BCL-1 MTC region. Sequence start at position 431 in the Genebank sequence with accession no. s77049. B. Sequence of BCL-1 mTC1p94 region. Reverse complement of accession no. ap001824 in the Genebank database. Sequence start at position 41535. C. Sequence of BCL-2 MBR region. Sequence start at position 2981 in the Genebank sequence with accession no. m14745 Citation[8]. D. Sequence of BCL-2 MCR region. Sequence from Ngan, 1989 Citation[27]. Sequence starts at position 905 and ends at position 1432.
![Figure 3. Breakpoints from the fusion DNA found in patients included in this study. Arrows indicate breakpoints and numbers refer to patient IDs. A. Sequence of BCL-1 MTC region. Sequence start at position 431 in the Genebank sequence with accession no. s77049. B. Sequence of BCL-1 mTC1p94 region. Reverse complement of accession no. ap001824 in the Genebank database. Sequence start at position 41535. C. Sequence of BCL-2 MBR region. Sequence start at position 2981 in the Genebank sequence with accession no. m14745 Citation[8]. D. Sequence of BCL-2 MCR region. Sequence from Ngan, 1989 Citation[27]. Sequence starts at position 905 and ends at position 1432.](/cms/asset/65a67a2d-6b48-4b35-b41f-b7373c669088/ionc_a_168125_f0003_b.gif)
Table III. PCR negative for t(11;14) and t(14;18). Patients and samples.