Figures & data
Figure 1. The timeline of HIF-1 research. The year-wise sequence of path breaking events in HIF-1 research are indicated with references. Events include the initial identification of HIF-1 up to the work being carried out in this field recently. The list is non-exhaustive and only a few major events are highlighted in the flowchart.
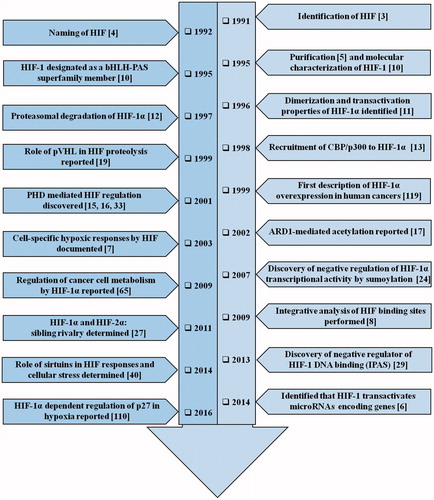
Figure 2. Structure and fate of the hypoxic master regulator. Figure presents the schematic representation of HIF-1α structure. N-terminal includes nuclear localization signal (NLS), followed by a basic helix-loop-helix (bHLH) domain that precedes two Per-ARNT-Sim homology domains (PAS-A and PAS-B). In addition to it, inhibitory domain (ID) separates N- and C-terminal transactivation domains (N-TAD and C-TAD). Figure also portrays the fate of HIF-1 through the sequence of events that occur during normoxic and hypoxic cellular microenvironment. During normoxia, certain post translational modifications lead to HIF-1α-pVHL binding and blocking of HIF-1α-p300/CBP interaction thereby causing HIF-1α degradation leading to the inhibition of HIF-1α-mediated transcription. During hypoxia, HIF-1α-pVHL binding does not occur, while p300/CBP can interact with HIF-1α, leading to transcriptional activation of target genes. Numbers highlights the amino-acid residues of various domains.
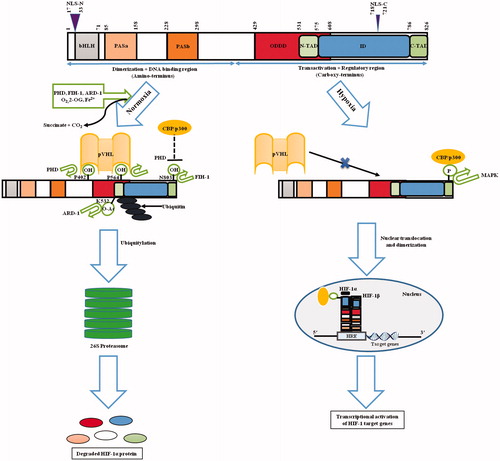
Figure 3. HIF-1 signaling cascade. Synthesis and constitutive expression of HIF-1α by a cascade involving a series of growth factors and signaling events are indicated. The major differences among the hypoxic and normoxic signaling and sequence of events is also depicted clearly in the flowchart. Normoxia leads to HIF-1α protein degradation whereas hypoxia leads to HIF-1α-regulated target gene expression. The downstream sequence of events leading to tumorigenesis is also portrayed.
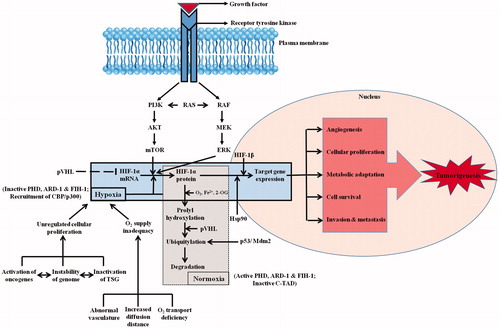
Table 1. Critical regulators of HIF-1 activity.
Figure 4. HIF-1 in the driver seat of tumorigenesis. The list of HIF-1 regulated genes (with corresponding references highlighted in bracket) having an impact on tumorigenic pathway is showcased. The HIF-1 target genes presented here are involved in processes such as cellular metabolism, proliferation and survival, angiogenesis, invasion and metastasis of tumor cells. The list is intended to be an illustration rather than being comprehensive.
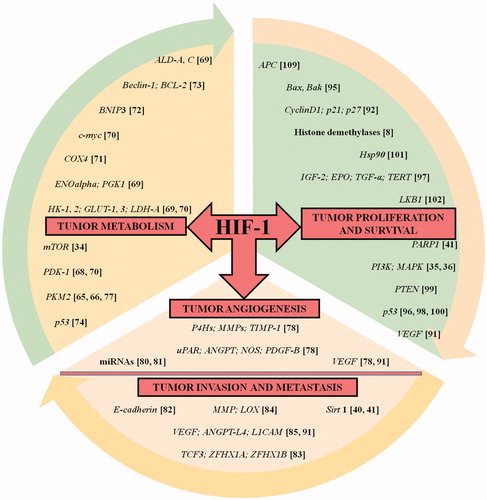
Table 2. Human cancers exhibiting increased levels of HIF-1α protein.
Table 3. Major chemical inhibitors of HIF-1 activity.
