Figures & data
Figure 1. Contrast-enhanced axial T1 MR image (A) and matching coronal T1 image (B) illustrating a right frontal glioblastoma with SVZ contact.
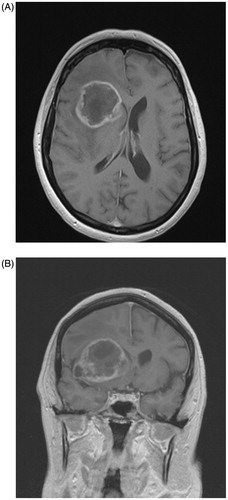
Table 1. Overview of demographic, surgical, molecular biological and volumetric characteristics as well as survival according to subventricular zone contact group.
Figure 2. Kaplan-Meier curves of overall survival (A) and progression-free survival (B) in glioblastoma with (SVZpos) and without (SVZneg) subventricular zone contact.
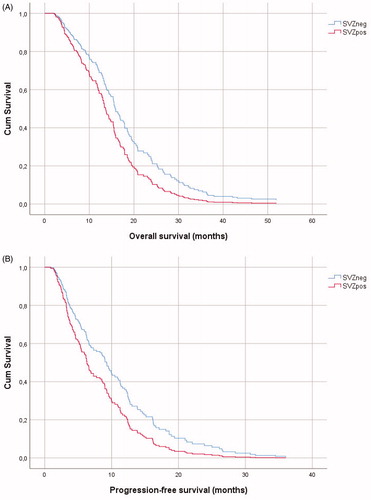
Table 2. Univariate survival analysis for several prognostic factors of glioblastoma patient survival.
Table 3. Multivariate Cox regression model of glioblastoma patient survival adjusted for age, KPS, extent of resection, MGMT-methylation and SVZ contact.
