Figures & data
Table 1. Patient characteristics at date of referral to the Phase 1 Unit.
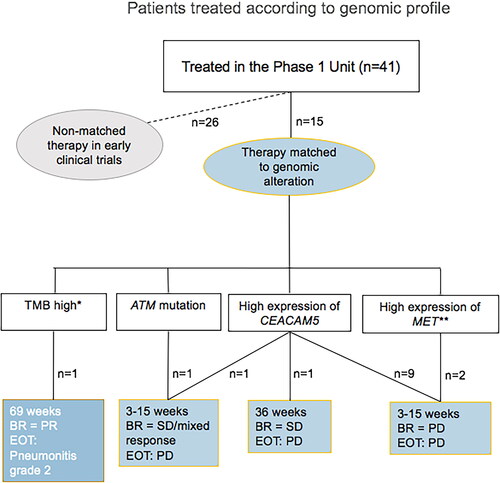
Figure 1. Pie charts of relative frequencies of different KRAS mutation subtypes among cohort. (a) All cancers. (b) Colorectal cancer (CRC). (c) Non-small cell lung cancer (NSCLC). (d) Pancreatic cancer. (e) All other cancers.
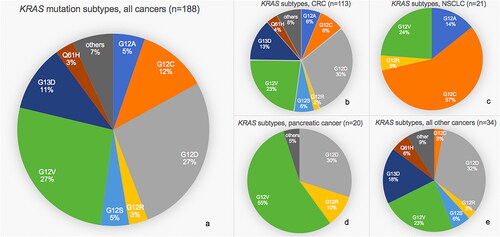
Figure 2. Bar chart of relative frequencies of co-occurring genomic alterations in different tumors. (a) In colorectal cancer (CRC), non-small cell lung cancer (NSCLC), pancreatic cancer, or all other cancers across all KRAS subtypes. (b) In tumors with KRAS mutation G12D, G12V, G12C, G13D, or all other subtypes across all cancers. Only alterations present in >5% of tumors are shown. Pathogenic: pathogenic mutation; vus: variant of unknown significance; HRD: homologous recombination deficiency; TMB: tumor mutational burden. PTEN loss include PTEN biallelic deletions and PTEN pathogenic mutation+PTEN deletion and/or loss of heterozygosity.
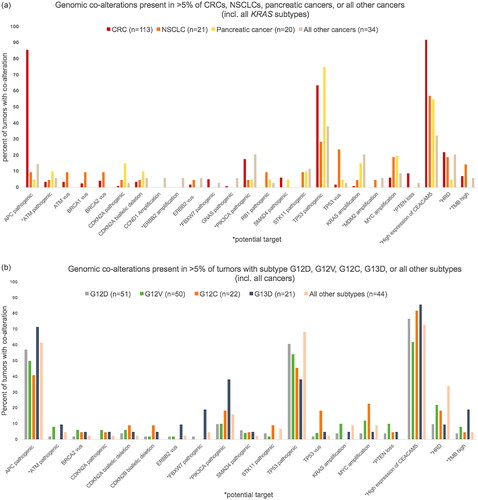
Figure 3. Prevalence of potential targets in 85 tumors with targets other than high expression of CEACAM5. *The patient with breast cancer who received BRAF targeted therapy. **With/without pathogenic mutations in mismatch repair genes. 62 tumors had one target, 17 tumors had two, five tumors had three and one tumor had four targets. HRD: homologous recombination deficiency; TMB: tumor mutational burden. PTEN loss include PTEN biallelic deletions and PTEN pathogenic mutation+PTEN deletion and/or loss of heterozygosity.
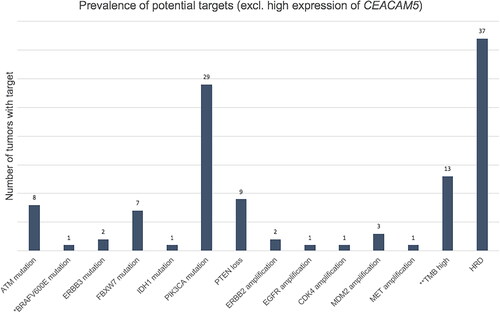
Figure 4. Patients treated according to genomic profile. *The patient received pembrolizumab based on high tumor mutation burden (TMB high). **Considered an actionable target at time of treatment. BR: best response according to RECIST 1.1; SD: stable disease; PR: partial response; PD: progressive disease; EOT: reason for end of treatment.
Supplemental Material
Download PDF (527.1 KB)Supplemental Material
Download MS Word (228.3 KB)Data availability statement
Data that underlie the results reported in this article can be made available upon reasonable request to the corresponding author, and will require the completion of a data processing agreement.
