Figures & data
Figure 1. Pathogenic variants identified in Sweden, plotted along the BRCA1-associated protein-1 (BAP1) gene with the functional domains shown. Ubiquitin carboxyl hydrolase (UCH) domain; HBM, host cell factor 1 (HCF1) binding domain; nuclear localization signals (NLS); C-terminal domain (CTD), additional sex combs like (ASXL1/2) binding domain; BRCA1-associated RING domain protein 1 (BARD1) binding region; Breast Cancer type 1 (BRCA1)-binding region and Ying Yang 1 (YY1) binding region; Forkhead Box Protein K1/2. 1American College of Medical Genetics (ACMG) evidence: PVS1 - null variant (nonsense, frameshift, canonical ±1 or 2 splice sites, initiation codon, single or multi-exon deletion) in a gene where LOF is a known mechanism of disease. PM1-mutational hotspot, PM2–Absent from controls in Exome Aggregation Consortium (ExAC). PM4–Protein length changes as a result of in-frame deletions/insertions in a nonrepeat region or stop-loss variants. PP1–Cosegregation with disease in multiple affected family members in a gene definitively known to cause the disease. PP3–Multiple lines of computational evidence support a deleterious effect on the gene or gene product (conservation, evolutionary, splicing impact, etc.). PP4–Patient’s phenotype or family history is highly specific for a disease with a single genetic etiology. PS3–Functional studies. 2Functional studies of the missense variant c.281A > G, demonstrated an almost complete abolishment of enzymatic activity (Repo et al. Hum Mol Genet. 2019). This together with a co-segregation of a highly specific phenotype, absence of variant in controls, computational evidence and position in a hotspot gene site gives evidence that the variant is likely pathogenic.
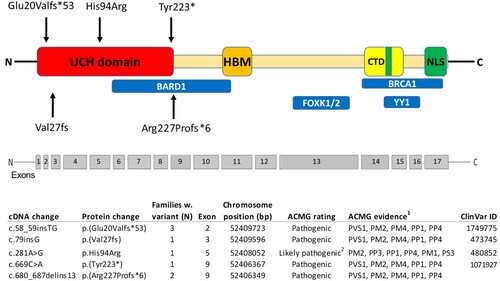
Table 1. BAP1 germline genetic testing in 190 families from Sweden.
Table 2. Tumors in BAP1 pathogenic variant carrying families identified in Sweden.
Supplemental Material
Download MS Word (14.5 KB)Data availability statement
The data that support the findings of this study are available from the corresponding author, upon reasonable request.
