Figures & data
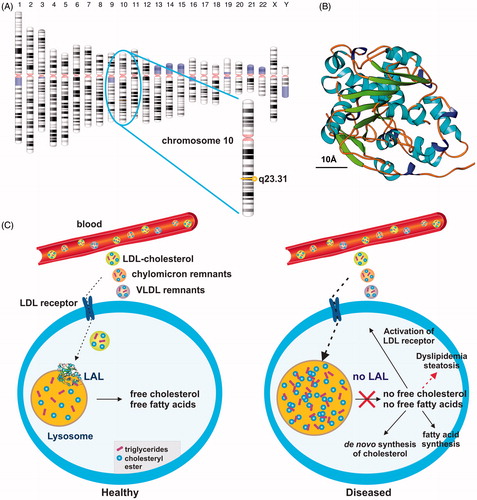
Figure 1. Simplified scheme of lysosomal acid lipase activity in cellular fat and cholesterol metabolism. (a) Lysosomal acid lipase is located on human chromosome 10q23.31. (b) Human LAL bears the characteristic structural features of the α/β hydrolase fold family consisting of typical α-helixes and β-sheetsCitation17. The predicted model of human LAL was generated using the fully automated protein structure homology-modeling server SWISS MODELCitation18, human LAL protein sequence deposited in the NCBI Protein data baseCitation19 (access. no. GenBank: AAB60328.1), and the crystal structure coordinates of dog gastric lipaseCitation20, sharing an overall sequence identity of 60.22% with human LAL. The estimated global model quality (GMQE) and global and local model quality (QMEAN) of the model are 0.77 and –2.99, respectively. A size marker (10 Å) is given. According to a Prosite scanCitation21, the active serine site is located at position 174 within the human LAL protein, consisting of a total of 399 amino acids. (c) In healthy subjects, LAL is located in lysosomes where it plays an important role in breaking down cholesteryl esters and triglycerides imported via the LDL/LDL receptor route from the blood into the cell. (d) The lack of LAL prevents breakdown of fatty material and trafficking of lipids from the lysosome into the cytoplasm. As a consequence, the control circuit for regulation of the intracellular cholesterol concentration is interrupted. In turn, the lower cellular cholesterol level provokes increased LDL receptor activity and stimulates cholesterol synthesisCitation22,Citation23. As a consequence, the cell is overloaded with cholesterol. Some of the extra cholesterol is a source for bile acid synthesis, while another portion becomes esterified and incorporated into VLDL, which ultimately can become trapped in the lysosomal compartment.