Figures & data
Figure 1. Damage caused by grass grub. A, Grass grub damage manifesting as dead patches in pasture (Canterbury). B, uplifted sward revealing high C. giveni larval densities with larvae forming chambers as depicted in C.

Figure 2. Photographs of grass grub and mānuka beetle larvae. A, C. giveni. B, P. festiva. Scale bar 5 mm.

Figure 3. Photographs of adult grass grub and mānuka beetle feeding on foliage. A, Nocturnal feeding by C. giveni during Spring flights. B, C. giveni feeding on deciduous leaves during Spring flights; C-D, P. festiva adult on mānuka (Leptospermum scoparium) during mating flights. Scale bar 10 mm.

Figure 4. Overview of the S. entomophila – C. giveni system. (A), Healthy C. giveni larvae (Scale bar 5 mm) ingest the roots (B) and associated S. entomophila. Within 2–5 days the larvae cease feeding, a result of as few as 500 anti-feeding prophage (Afp) particles. The proposed model for the action of the Afp is depicted in (C), where the extended virus-like Afp particle latches onto a currently undetermined receptor on cuticular membranes or the intestinal cells using its tail fibres. Once bound (cell attachment), the Afp particle changes conformation to inject the Afp loaded toxin into the eukaryotic target cell. The S. entomophila pathogenicity (Sep) Toxin Complex (Tc) (D) is released by ingested bacteria. This causes gut clearance, expulsion of gut contents (E) and the larvae to become amber in colour (F); scale bar 5 mm. The disease process can last for more than 4 months. There are no apparent sites of S. entomophila colonisation on the surface of midgut cells, with bacteria only adhering to particulate matter of the gut and membranous surfaces (G). Within 6 days of ingestion, bacteria multiply within the fermentation chamber of the insect (H). Under conditions of high cell density levels of the S. entomophila-derived chitinases are reduced (I), possibly prolonging the time of infection. I, measurements of chitinase activity; WT, denotes wildtype S entomophila; Mutant, denotes quorum sensing (QS) mutant; +AHL denotes the mutant complemented by addition of an exogenous supply of the QS signal molecule N-Acylhomoserine lactone (AHL). Figure modified from Hurst (Citation2016).
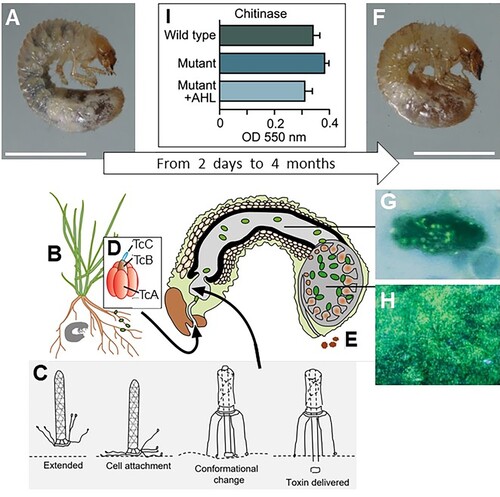
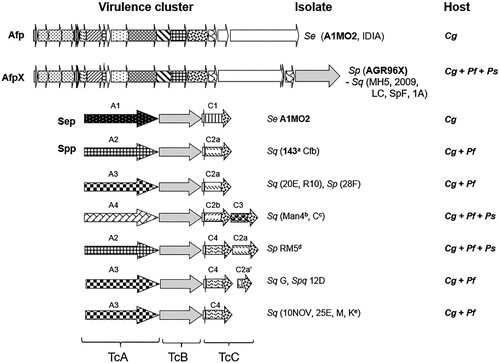
Figure 5. Schematic of the S. entomophila (Se), S. proteamaculans (Sp) and S. quinivorans (Sq) Afp and Tc variants and their associated C. giveni (Cg), P. festiva (Pf), and P. setosa (Ps) host. Patterned arrows de note the Tc (TcA1-A4, TcC1 – C4) variants. Relevant isolate identifiers are listed where bold text denotes the previously documented S. entomophila Sep and Afp and the S. quinivorans isolate 143, and AGR96X AfpX virulence-associated region. Isolate names are listed where superscript letters indicate: ano activity against Pyronota species, btransient activity against C. giveni, cno activity against C. giveni, transient activity against Pyronota species, dtransient activity against Pyronota spp., e Pyronota-active. Figure modified from Hurst et al. (Citation2021).