Figures & data
Table 1 Location, land tenure, rainfall for study sites and treatments, and sampling dates.
Figure 1 Total density (n/m2) of the 15 most speciose families from each site shown for each treatment in order of abundance in the native tussock treatments.
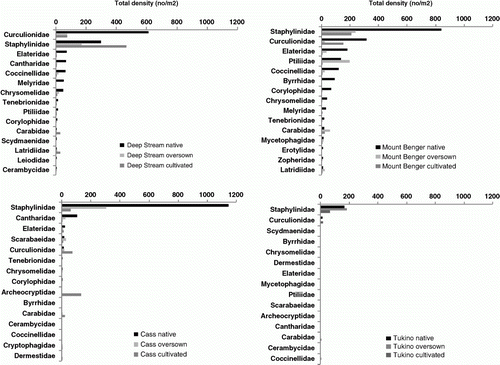
Figure 2 Number of species in the 15 most speciose families from each site shown for each treatment plotted in order of abundance in the native tussock treatments.
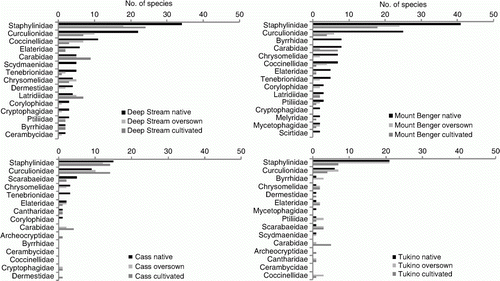
Table 2 Summary of Coleoptera families, species richness (S), density, total and exotic species found at each site and vegetation type, and percentage of species in three main trophic groups.
Figure 3 Coleoptera species accumulation curves for Deep Stream and Mount Benger, showing 95% confidence limits (fine solid and dashed lines).
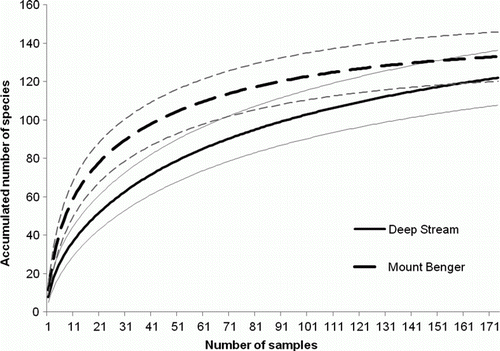
Table 3 Species diversity indices for each site and treatment.
Figure 6 Values for distance from lognormal fitted for A, mean of all sites excluding Cass for each vegetation treatment; B, each site and treatment. The closer χ2/n is to zero, the better the fit to a lognormal distribution, and hence the least disturbed as indicated by the distribution of Coleoptera species abundance across species.
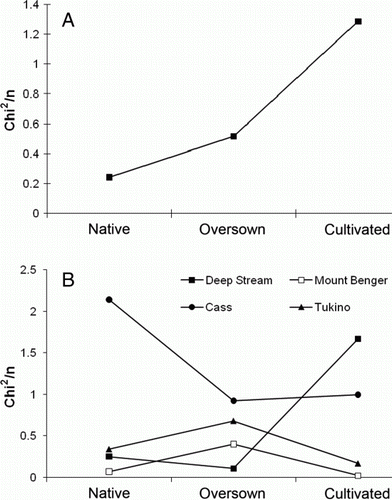
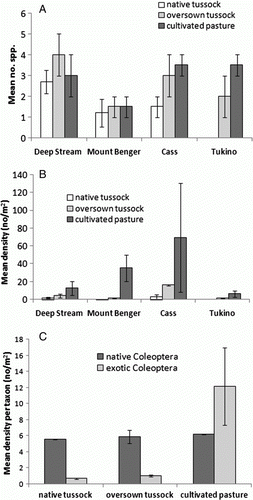
Figure 7 Mean number (±SE) A, and density (±SE) B, of exotic Coleoptera species in the three vegetation types at each locality; mean density per taxon (±SE) C, of native and exotic Coleoptera in each vegetation type for all four sites.