Figures & data
Figure 1 A, Distribution of Cyanoramphus parakeets in New Zealand and the sub-Antarctic based on modern and historical records. See key for distributions of: yellow-crowned parakeet (Cyanoramphus auriceps); orange-fronted parakeet (Cyanoramphus malherbi); red-crowned parakeet (Cyanoramphus novaezelandiae novaezelandiae); Chatham Islands red-crowned parakeet (Cyanoramphus n. chathamensis); Antipodes Island parakeet (Cyanoramphus unicolor); Reischek's parakeet (Cyanoramphus hochstetteri); and Forbes parakeet (Cyanoramphus forbesi). The red-crowned parakeet (C. n. novaezelandiae) at Campbell Island and the Macquarie Island parakeet Cyanoramphus n. erythrotis (Wagler, 1832) are extinct (Chambers & Boon Citation2005; Scofield Citation2005; Holdaway et al. Citation2010). B, Distribution of red-crowned, yellow-crowned and hybrid (the hybrids are red-crowned morph/orange-fronted haplotype and yellow-crowned morph/orange-fronted haplotype) parakeet DNA samples from the Auckland Islands. Numbers indicate sample sizes. (For a colour version of this figure, the reader is referred to the online version of this article.)
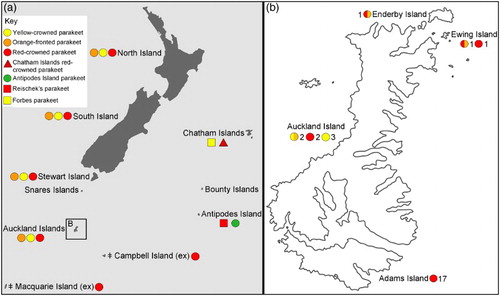
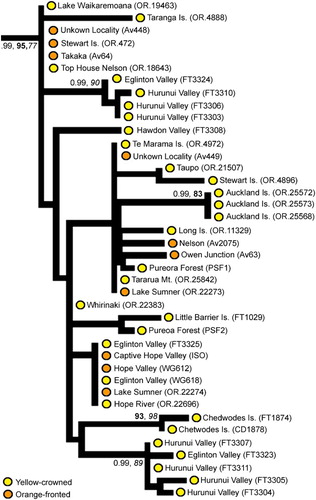
Figure 2 Phylogeny of Cyanoramphus parakeets based on 1605 bp mitochondrial DNA CR alignment. Branch lengths are proportional to the number of substitutions/site. The maximum likelihood tree was generated in PhyML using the GTR + I + G model of nucleotide substitution. For clarity, only Bayesian posterior probability (PP) and bootstrap support (BS; bold = maximum likelihood; italics = maximum parsimony) values for major clades are shown. The asterisks indicate strong support from all of these measures, i.e. Bayesian posterior probability of 0.95 and above, and bootstrap values of 80% and above. Coloured bars represent the dominant crown colouration of each species as per . Clades in grey-outlined boxes are shown in detail in (Cyanoramphus novaezelandiae and Cyanoramphus malherbi) and (Cyanoramphus auriceps). (For a colour version of this figure, the reader is referred to the online version of this article.)
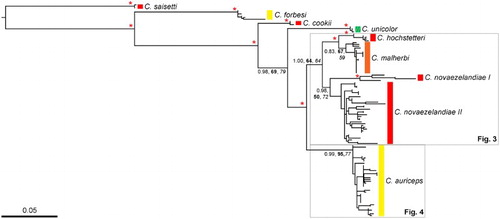
Figure 3 Sub-phylogeny of red-crowned (Cyanoramphus novaezelandiae) and orange-fronted (Cyanoramphus malherbi) parakeets based on 1605 bp mitochondrial DNA CR alignment, showing the phylogenetic position of red-crowned and orange-fronted parakeet lineages from the Auckland Islands (from Adams, Enderby, Ewing and Auckland Islands), and orange-fronted parakeets from mainland New Zealand. For clarity, for internal nodes only Bayesian posterior probability (PP, > 0.95) and bootstrap support (bold = maximum likelihood; italics = maximum parsimony, > 80%) values for major clades are shown where the node has strong support from at least two different methods. The asterisks indicate strong support from all of these measures, i.e. Bayesian posterior probability of 0.95 and above, and bootstrap values of 80% and above. Lineage colours represent the dominant crown colouration of each species, while coloured circles represent the morphotype of individual specimens. (For a colour version of this figure, the reader is referred to the online version of this article.)
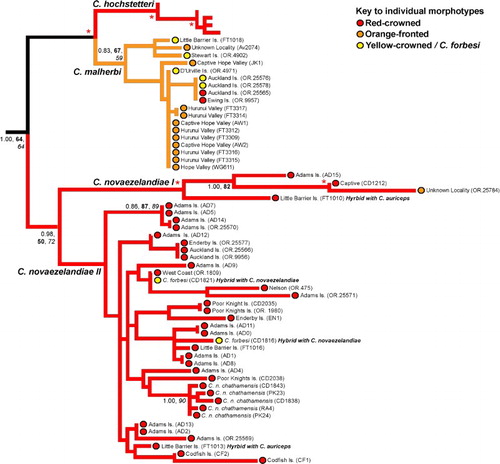
Figure 4 Sub-phylogeny of yellow-crowned parakeets (Cyanoramphus auriceps) based on the 1605 bp mitochondrial DNA CR alignment showing the position of yellow-crowned parakeets from the Auckland Islands. For clarity, for internal nodes only Bayesian posterior probability (PP, > 0.95) and bootstrap support (bold = maximum likelihood; italics = maximum parsimony, > 80%) values for major clades are shown where the node has strong support from at least two different methods. Yellow-crowned and orange-fronted morphotypes have been indicated on the phylogeny. (For a colour version of this figure, the reader is referred to the online version of this article.)