Figures & data
Figure 1. Amplification of the σC-encoding and σNS-encoding genes of ARV isolates. The PCR product (1022 bp) was amplified from the σC-encoding gene of ARV isolates using primer pairs S1A/S1H (1a) and S1A1/ S1B1 (1b). The PCR product (1152 bp) was amplified from the σNS-encoding gene of ARV isolates using primers S41 and S42 (c). Lane M, molecular weight marker of 1 kb DNA ladder; lane 1, S1133; lane 2, 2408; lane 3, 750505; lane 4, 601SI; lane 5, 601G; lane 6, R2/TW; lane 7, 916; lane 8, 918; lane 9, 1017-1; lane C, negative control.
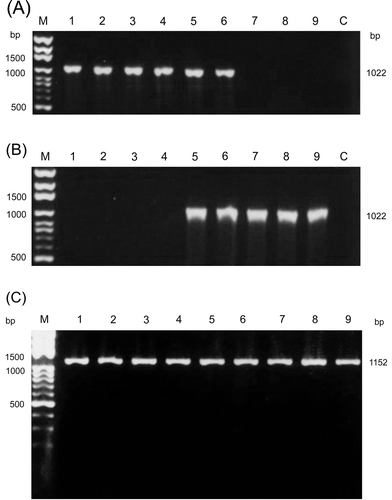
Figure 2. Polyacrylamide gel electrophoretic patterns of the PCR product (1022 bp) from the σC-encoding gene of each isolates digested with restriction enzymes Bcn I (1a), Hae III (1b), Taq I (1c), Dde I (1d), and Hinc II (1e). Lane M, molecular weight marker of 1 kb DNA ladder; lane 1, S1133; lane 2, 2408; lane 3, 750505; lane 4, 601SI; lane 5, 601G; lane 6, R2/TW; lane 7, 916; lane 8, 918; lane 9, 1017-1 The arrows indicate the faint bands. A diagram showing the location of restriction siteis is shown in .
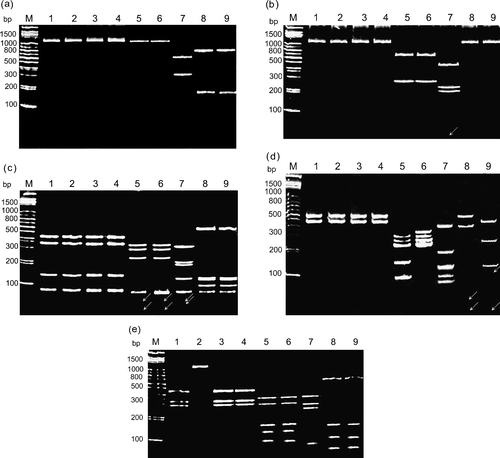
Figure 3. Diagram of restriction enzyme digestions of the σC-encoding gene of ARV isolates. The genome segment S1 of ARV S1133 is indicated by the box. The amplified cDNA fragment (1022 bp) is numbered from 601 to 1622. The restriction sites of the amplified cDNA fragments from each strain are shown as Bcn I (B), Dde I (D), Hae III (Ha), Hinc II (H), and Taq I (T). The classification of ARV isolates is indicated one the right-hand side.
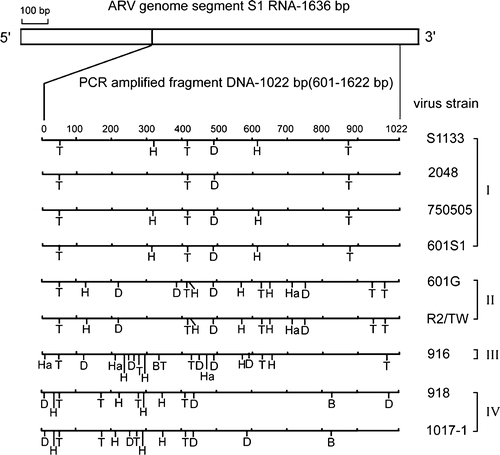
Figure 4. Polyacrylamide gel electrophoretic patterns of the PCR product (1152 bp) from the σNS-encoding gene of each isolate digested with restriction enzymes Bcn I (4a), Hae III (4b), and Taq I (4c). Lane M, molecular weight marker of 1 kb DNA ladder; lane 1, S1133; lane 2, 2408; lane 3, 750505; lane 4, 601SI; lane 5, 601G; lane 6, R2/TW; lane 7, 916; lane 8, 918; lane 9, 1017-1. The arrows indicate the faint bands. A diagram showing the location of restriction site is shown in .
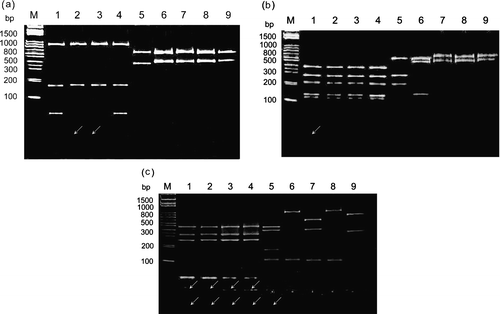
Figure 5. Diagram of restriction enzyme digestions of the σNS-encoding gene of ARV isolates. The genome segment S4 of ARV S1133 is indicated by the box. The amplified cDNA fragment (1152 bp) is numbered from 6 to 157. The restriction sites of the amplified cDNA fragments from each strain are shown as Bcn I (B), Dde I (D), Hae III (Ha), Hinc II (H), and Taq I (T).
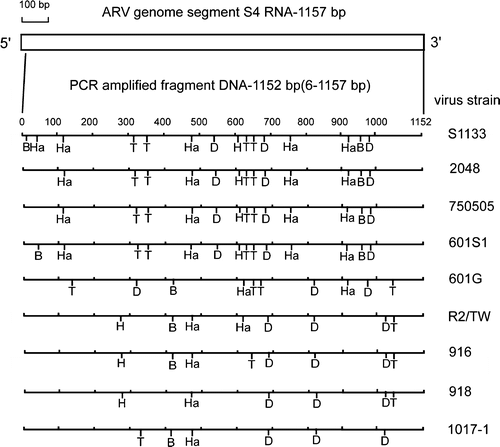
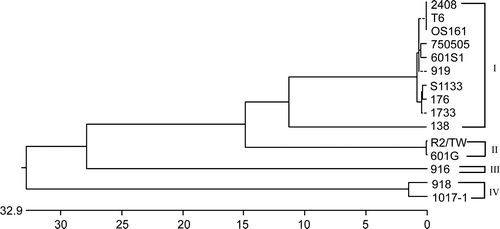
Figure 6. A phylogenetic tree based on nucleotide sequences of the σC-encoding gene (1022 bp). The ARV isolates in each group are indicated on the right-hand side.