Figures & data
Figure 1. Degranulation of heterophils isolated from 2-day-old chickens was compared to determine their ability to release toxic granules when stimulated with opsonized S. enteritidis. Data are pooled and presented as the mean±standard error of the mean of the µM β-d glucuronidase released from triplicate assays from three separate experiments (n = 450 chickens). All statistical differences are for comparisons between females (FF vs SF) or between males (FM vs SM) (*P < 0.05).
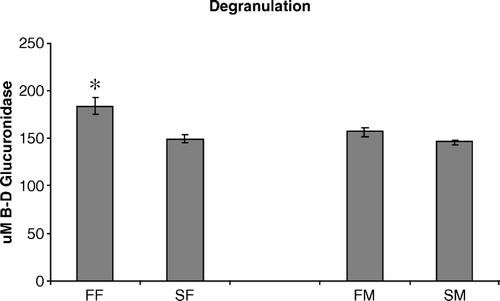
Figure 2. Heterophils isolated from FF, SF, FM, and SM 2-day-old chickens were compared to determine their ability to generate an oxidative burst response following stimulation with the inflammatory agonist phorbol A-myristate 13-acetate. Data are pooled and presented as the mean±standard error of the mean of the RFU of triplicate assays from three separate experiments (n = 450 chickens). All statistical differences are for comparisons between females (FF vs SF) or between males (FM vs SM) (*P < 0.05).
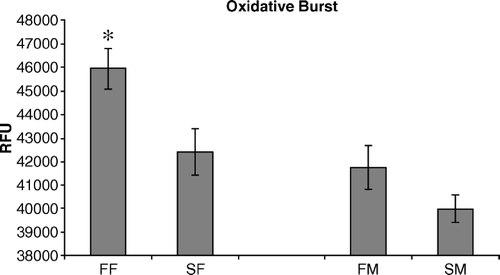
Table 1. Control, S. enteritidis-treated, and fold change of IL-6, CXCLi2, and IFN-α mRNA expression levels in heterophils isolated from F1 cross C and D chickens separated by sexa
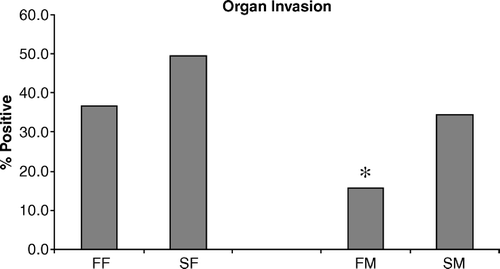
Figure 3. Two-day-old chickens were challenged orally with S. enteritidis (5×106 CFU/chicken) and liver/spleen organ invasion was determined using standard microbiological techniques. Data are pooled from 90 chickens and presented as the mean±standard error of the mean of triplicate organ invasion experiments. Data shown are the percentages obtained from the number of S. enteritidis-positive chickens/90. All comparisons are between females (FF vs SF) or between males (FM vs SM) (*P < 0.05).