Figures & data
Table 1. Characterization of R. anatipestifer isolates by seven different methods
Figure 1. Cluster analysis of 24 R. anatipestifer isolates constructed using ClustalV to analyse nucleotide sequences of ompA. Bootstrap values were estimated for this tree using the neighbour-joining model for 1000 replicates. Arabic number, bootstrap value of each cluster.
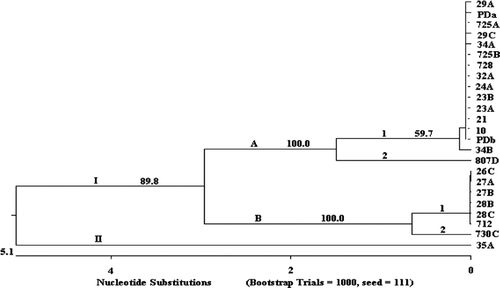
Figure 2. Nucleotide sequence alignment and comparison of the ompA of five representative R. anatipestifer isolates. Underlined region, restriction site of restriction enzyme AluI. Arrow, direct repeat region. Repeats II and III encode for PEST regions (Subramaniam et al., Citation2000), and two are imperfect repeats with a sequence diverse from repeats II and III. –, deletion.
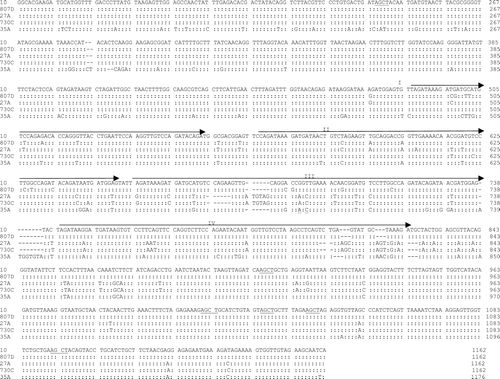
Figure 3. Rep-PCR patterns of 24 R. anatipestifer isolates. M, 1 kb size marker ranging from 200 bp to 14 kb. N, Negative control (PCR mixture without DNA template).
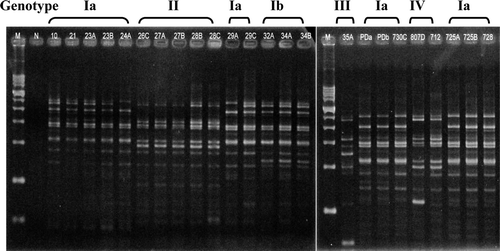
Figure 4. PFGE patterns of R. anatipestifer isolates. Genomic DNAs were digested by restriction endonuclease SmaI. M, DNA size marker (48.5-kb phage ladder; BIO-RAD, USA).The same Arabic number means R. anatipestifer isolates were isolated from the same goose or duck. Different letters indicate R. anatipestifer isolates were isolated from different tissues.
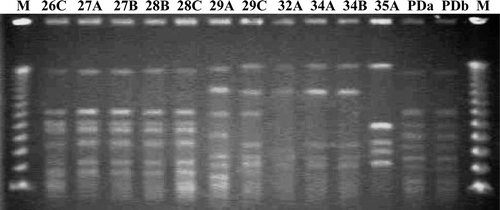
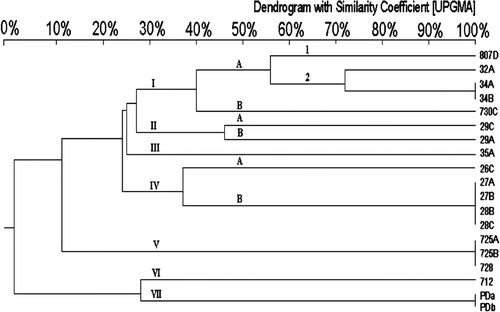
Figure 5. Dendrogram of 24 R. anatipestifer isolates constructed by UPGMA with 3% tolerance to analyse SmaI-digested PFGE pattern. The same Arabic number means R. anatipestifer isolates were isolated from the same goose or duck. Different letters indicate R. anatipestifer isolates were isolated from different tissues.