Figures & data
Table 1. Type of samples processed and the results of virus isolation and RRT-PCR from different clinical samples collected in Houbara bustards and falcons
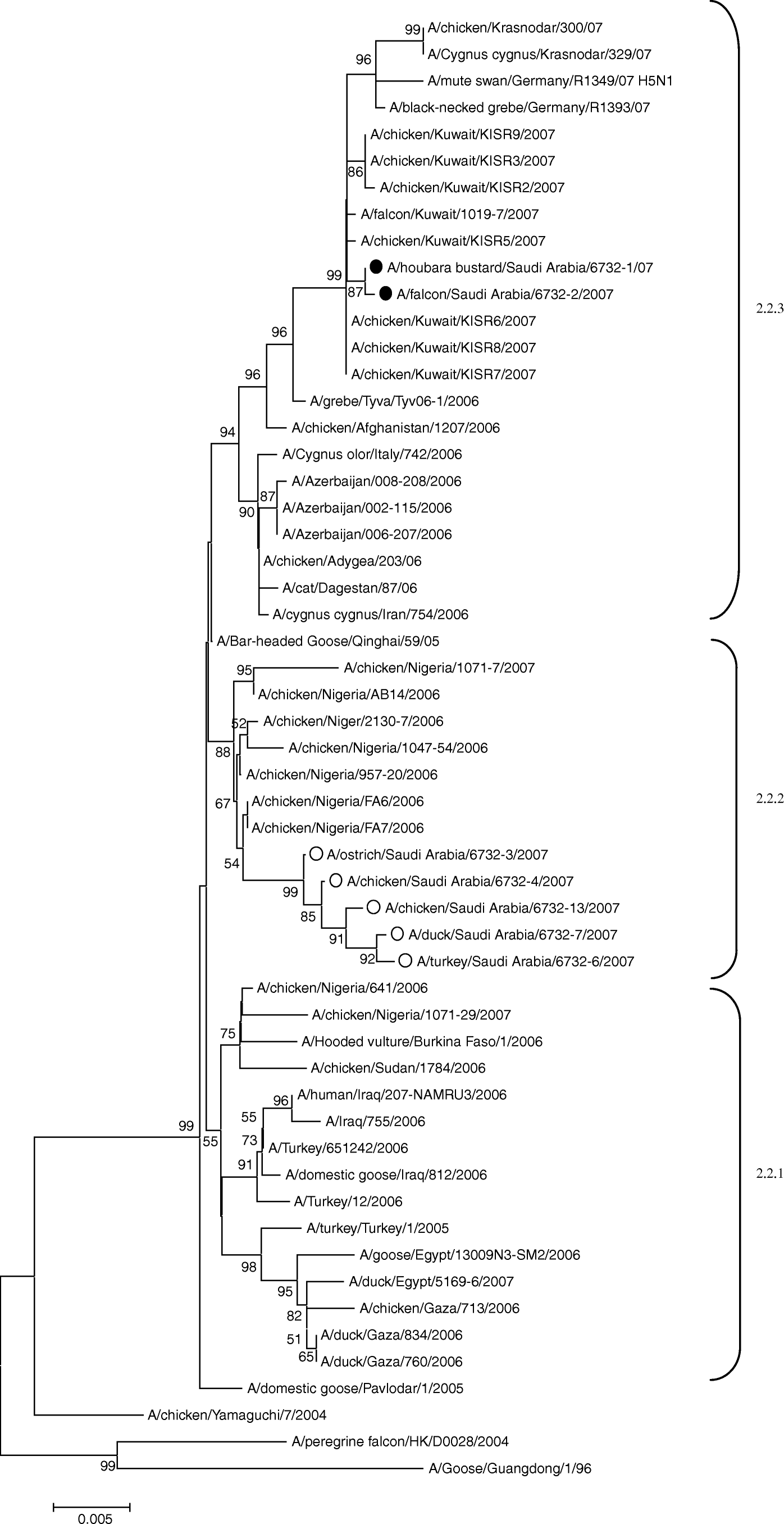
Figure 1. Unrooted phylogenetic tree for representative HA nucleotide sequences of clade 2.2 constructed by the neighbour-joining method. Black circles, sequences generated from the viruses described in this study. Selected A/H5N1 viral sequences are included for comparison. White circles, sequences generated from viruses isolated in poultry in the KSA. Bootstrap values >50% are shown at the nodes. Genetic sublineages included in lineage 2.2 are indicated on the right-hand side.