Figures & data
Figure 1. Map of China showing Shanghai and Shandong, Henan, Anhui, Jiangsu, Zhejiang and Fujian provinces, where sampling ducks came from during the Newcastle disease virus surveillance program at LBMs.
Table 1. NDV isolates from the surveillance programme in domestic ducks at LBMs in Eastern China during 2002 to 2007 used for biological and genetic characterization
Table 2. Sequences of oligonucleotide primers used in RT-PCR assays for amplification of F genes of NDV isolates
Table 3. Background information for representative NDV strains including previously established class II genotype I to genotype IX and the more recently defined class I genotype 1 to genotype 9 viruses used as reference in this study
Table 4. Monthly isolation of NDVs from cloacal swab specimens of domestic ducks at wholesale live bird markets in Eastern China during July 2002 to June 2007
Figure 2. Phylogenetic tree of NDV reference strains representing previously established class II genotype I to IX and the more recently set up class I genotype 1 to 9, and representative NDV isolates from surveillance in domestic ducks in this study (set in bold), showing major divisions of classes and genotypes. The tree is based on comparison of partial F gene sequence (between nt 47 and 420). Accession number and other background information of the reference strains are in .
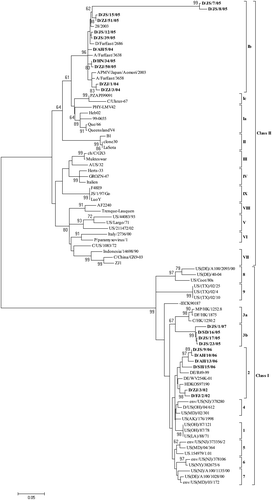
Figure 3. Phylogenetic tree of NDV reference strains representing the more recently established class I genotypes 1 to 9 viruses, and all the 30 class I isolates from surveillance in domestic ducks in this study (set in bold), showing major genotypes and their relationships in this class. The tree is based on comparison of partial F gene sequence (between nt 47 and 420). Accession number and other background information of the reference strains are in .
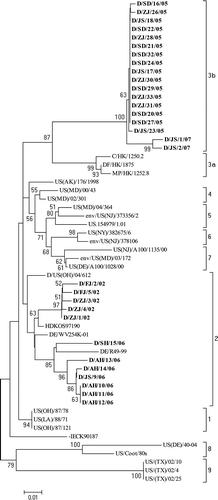
Figure 4. Phylogenetic tree of NDV reference strains representing the previously established class II genotypes I to IX viruses, and all the 43 class II isolates from surveillance in domestic ducks in this study (set in bold), showing major genotypes and their relationships in this class. The tree is based on comparison of partial F gene sequence (between nt 47 and 420). Accession number and other background information of the reference strains are in .
