Figures & data
Table 1. Characterization of NDV strains used in the present study.
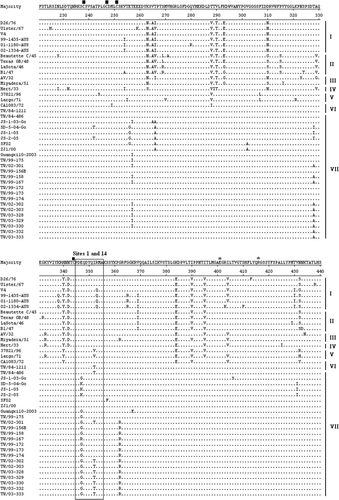
Figure 1. HN amino acid sequences were deduced from the HN nucleotide sequence of NDV. Alignment of HN amino acid sequences of recent Taiwanese isolates and previously published virus strains was performed using the DNASTAR software. Amino acid residues of HN protein of different NDV strains are shown in a single-letter code. Residues that are identical to the majority are indicated by a dot (.). The HRA and HRB (amino acid residues 74 to 110) and C-terminal extension are indicated above the amino acid sequences. Variation at amino acid residue 81 in HRA is indicated above amino acid sequences. A total of 13 cysteine residues in the HN glycoprotein of NDV strains, at residues 123, 172, 186, 196, 238, 247, 251, 344, 455, 461, 465, 531, and 542, are marked by ▪ above the amino acid sequences. Three domains related to epitopes on HN protein of NDV, including site 23, sites 1 and 14, a C-terminal domain composed of residues 494, 513 to 521, and sites 12 and 2, are indicated by boxes. Three key amino acid residues at positions 401 (E), 416 (R), and 526 (Y) of HN glycoprotein required for receptor binding are indicated by * above the amino acid sequences. Phylogenetic lineages created on the basis the F gene of NDV are indicated on the left-hand side. The majority indicates the common HN sequences of NDV.
Table 2. HA titre and amino acid sequences at residue 81 in HRA of NDV strains.
Figure 2. 2a: Real-time RT-PCR fluorescence curve derived from serially diluted NDV genomic RNA. Following amplification, the real-time RT-PCR products were analyzed by agarose gel electrophoresis. 2b: Standard curve of real-time PCR. Serially diluted NDV genomic RNA was amplified and analyzed in real time. The threshold cycle (Ct) values were plotted against the copy number to construct the standard curve, r=–1.0. The NDV RNA copy number was determined spectrophotometrically.
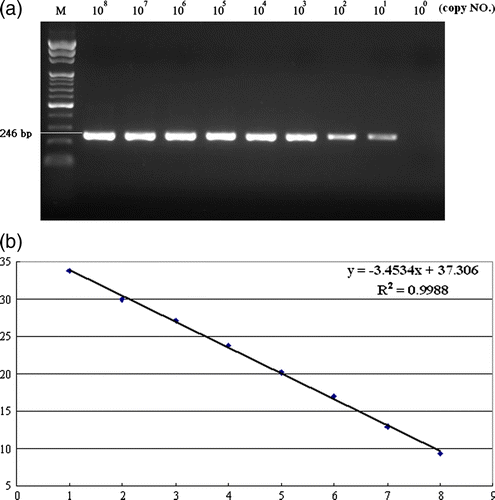
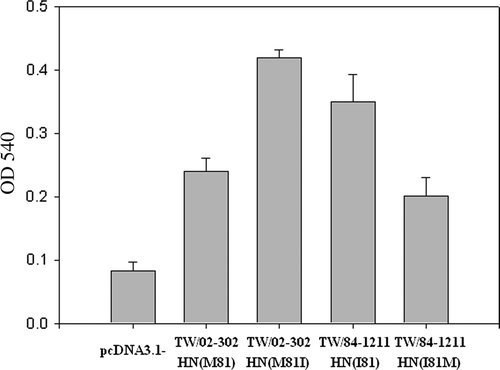
Figure 3. Haemadsorption activity of HN mutants was determined at 4°C by the ability of the expressed protein to adsorb chicken red blood cells. Two mutants, including TW/02-302 HN M81I and TW/84-1211 HN I81M created from NDV TW/02-302 HN M81 and TW/84-1211 HN I81 strains, are shown. The HN mutants (TW/02-302 HN M81I and TW/84-1211 HN I81M) showed significantly increased or reduced haemadsorption activity (P<0.05) when compared with TW/02-302 HN M81 and TW/84-1211 HN I81 strains. Haemadsorption activity was quantitated by measurement of the absorbance at 540 nm minus the background obtained with cells expressing the vector alone. The results shown represent the means of three independent experiments. Error bars represent the mean±standard deviation.