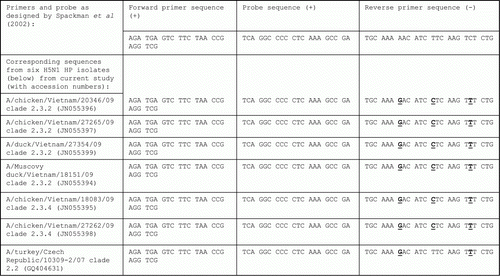
Figures & data
Table 1. Description of four H5N1 HPAI-infected Vietnamese poultry flocks sampled in 2009.
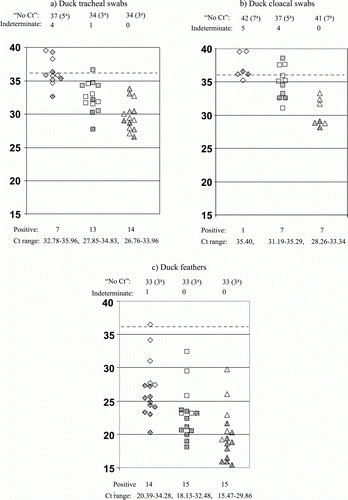
Figure 1. Distribution of Ct values for different types of clinical specimens from chickens tested by both M-gene RRT-PCRs and the H5 HA2 RRT-PCR. Specimens from all 46 chickens included (a) tracheal swabs, (b) cloacal swabs and (c) feathers. Ct values (vertical axis) for M gene ‘wet’ and ‘bead’ RRT PCR results are shown by diamond and square symbols respectively, H5 HA2 RRT PCR Ct values are shown by triangles. Filled symbols indicate chicken specimens that were tested by VI, where dot and grey fills indicate VI positive and VI negative results respectively. The total numbers of “No Ct” results for each RRT PCR are stated above the corresponding test, with parentheses indicating numbers of VI negative and positive results by a and b respectively. Numbers of indeterminate RRT PCR results (Ct 36.01–39.99) are also stated above each graph (broken horizontal line indicates Ct 36), while numbers of positive RRT PCR results are stated beneath each graph for the corresponding test. The range of Ct values are listed for the positive results for the corresponding RRT PCR.
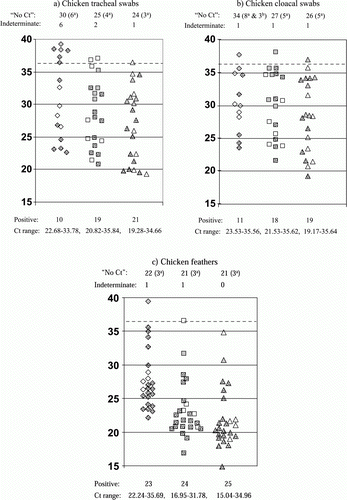
Figure 2. Distribution of Ct values for different types of clinical specimens from ducks tested by both M-gene RRT-PCRs and the H5 HA2 RRT-PCR. Specimens from all 48 ducks included (a) tracheal swabs, (b) cloacal swabs and (c) feathers. All were tested by the three RRT PCRs described in Fig 1, and details are as explained in the footnote to the preceding figure. Fig 2 includes VI results for 32 duck specimens.