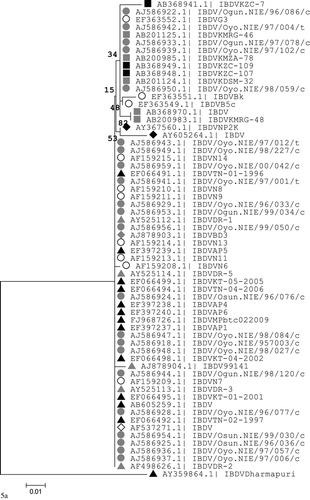
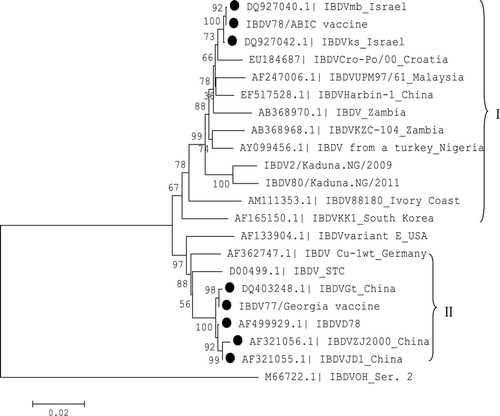
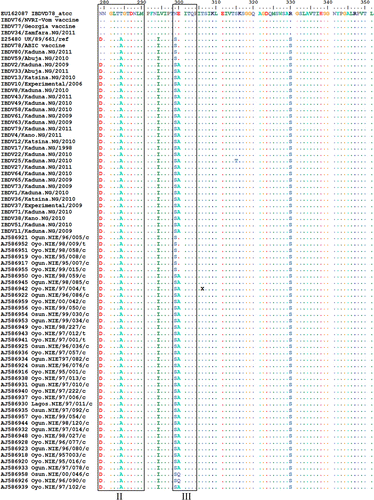
Figures & data
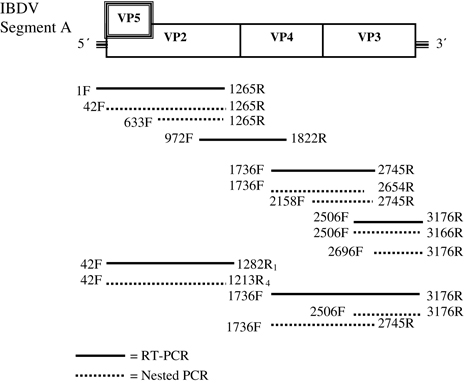
Table 1. Primers used to amplify and sequence IBDV VP2 and genome segment A.
Table 2. Origin of field and vaccine IBDV-positive samples as tested with PCR.
Table 3. Comparison of segment A amino acids for vaccine and field IBDV strains in northwestern Nigeria.
Table 4. Comparison of IBDV VP2 amino acid positions 299 and 300 as proposed regional markers for different countries.
Supplementary Table S1. Statistical test for association between proposed infectious bursal disease virus regional markers and different groups of countries