Figures & data
Table 1. Mortality in flocks of laying chickens with EPS from which E. coli colonies were subjected to PFGE and MLST genotyping.
Table 2. Results of gross post-mortem examination and bacteriology of dead hens originating from chicken flocks with EPS.
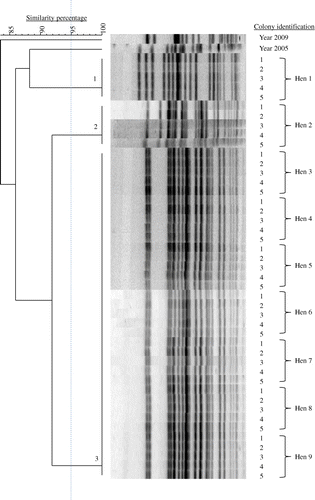
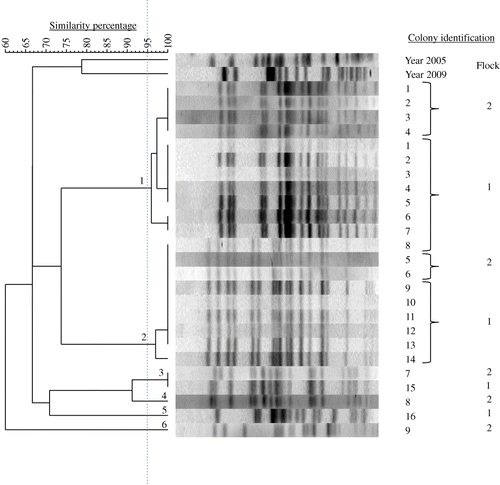
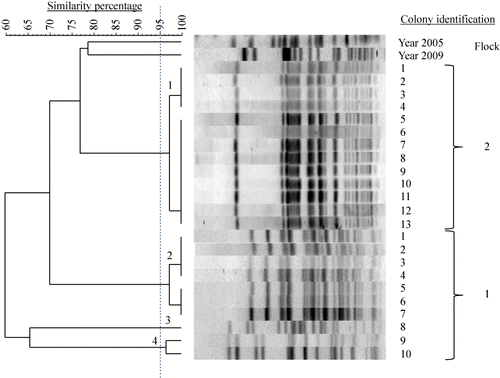
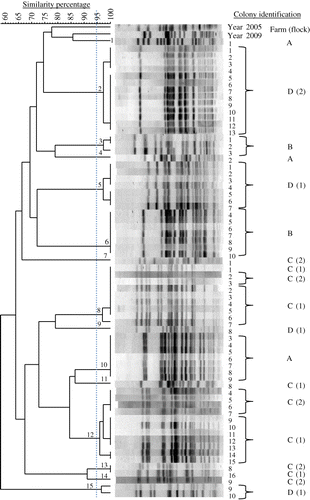
Table 3. Number and prevalence of E. coli PFGE genotypes in chicken flocks with EPS.
Table 4. Target genes, primer name, primer sequence and product size of the seven housekeeping genes used for multilocus sequence typing of E. coli (Wirth et al., Citation2006) isolated from the bone marrow of hens with EPS.