Figures & data
Table 1. Seventeen PPMV-1 strains characterized in this study.
Table 2. Identity matrix of F gene partial nucleotide (nucleotides 47–420) (lower triangle) and amino acid (upper triangle) sequences of pigeon-origin APMV-1 characterized in the present study. The sequence identity matrix was generated by BioEdit software (Hall, Citation1999).
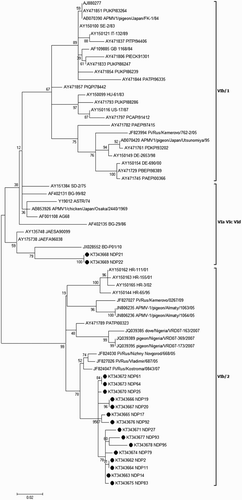
Figure 1. Phylogenetic tree based on the variable region (nucleotides 47–420) of the fusion protein gene representing different genotypes of class II, including reference strains, the Iranian strains (AY928937, AY928933, AY928934, JQ267579) and the isolates obtained in current study (bullet points). Genotype designation based on Diel et al. (Citation2012) is mentioned before accession numbers. Bootstrap values (500 replications) are shown next to the branches.
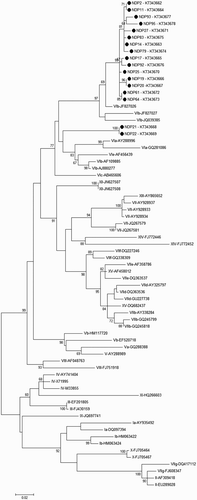
Figure 2. Phylogenetic analysis of 64 PPMV-1 strains based on the variable region (nucleotides 47–420) of the fusion protein gene. Bootstrap values (500 replications) are shown next to the branches. Genotype designation according to Ujvári et al. (Citation2003) and Diel et al. (Citation2012) is given to the right. Dots indicate isolates of the present study.