Figures & data
Figure 1. Gross lesions in gizzards from 41-day-old pullets (pullet flock P-A). Erosions in the koilin layer (arrows).
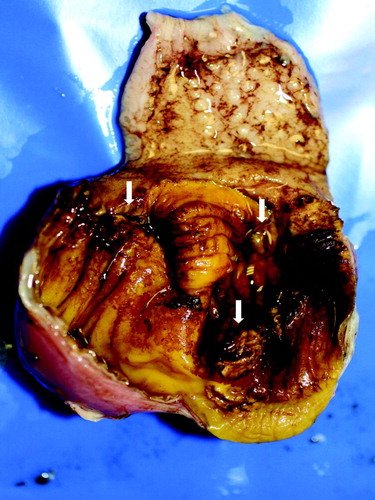
Table 1. Data on the pullet and layer flocks investigated together with results from histopathological and virological investigations of gizzards.
Figure 2. Weekly mortality (%) in (a) the affected pullet flock (P-A) and (b) the affected layer flocks (L-A and L-B - black and grey bars, respectively).
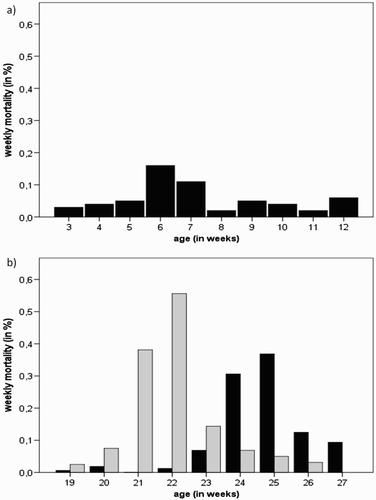
Figure 3. Histopathology of gizzards from 35-day-old pullets (pullet flock P3). (a) Erosion of the epithelium and infiltration of inflammatory cells in the mucosal membrane. Basophilic intranuclear inclusion bodies (arrows) can be observed in gizzard epithelial cells. H&E. Bar = 100 µm. (b) Inclusion bodies with FAdV-A DNA. ISH. Bar = 20 µm.
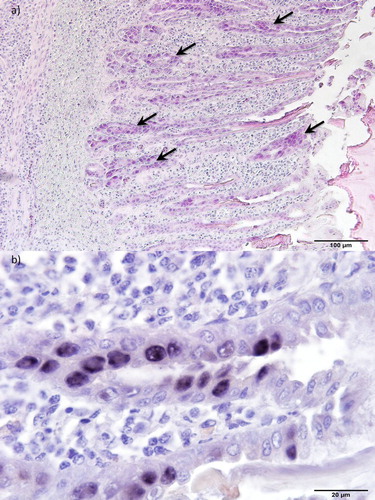
Figure 4. Box plot presentation of VNT results from pullet and layer cases. Titres (log2) of FAdV-1-, FAdV-4- and/or FAdV-8a-specific antibodies from pullet flocks with (P-A) and without gizzard erosions (P-B, “control”) and from a layer flock (L-B) collected at the time disease was diagnosed (P-A1, P-B1 & L-B1) and approximately six (P-A2, P-B2)/three (L-B2) weeks later are shown. Titres ≤ 3 log2 are considered negative. Different letters (a–d) indicate statistically significant differences (P < 0.05).
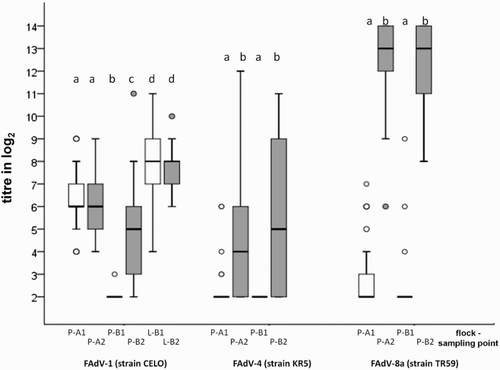
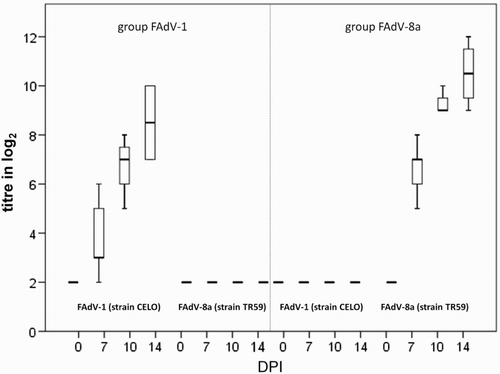
Figure 5. Box plot presentation of VNT results from in vivo study. Titres (log2) of FAdV-1- and FAdV-8a-specific antibodies from SPF pullets orally infected with FAdV-1 or FAdV-8a. Titres ≤ 3 log2 are considered negative.