Figures & data
Table 1. Strains, plasmids and primers used in this study.
Figure 1. Identification and characterization of mutant strain ΔsprA and complemented strain CΔsprA. Usually, a combined PCR method is used to verify the gene deletion strain. In this study, we determine whether homologous recombination occurred by amplifying spc gene. In terms of experimental design, the leftarm (1055 bp) and rightarm (1226 bp) of the constructed suicide plasmid were 5742 bp (containing a 121 bp fragment) shorter than the target sprA gene (7023 bp), which could be used as an internal control to determine whether the complete recombination occurred. If the gene deletion strain could not amplify this 121 bp fragment, it indicated that the gene deletion strain was successfully constructed; otherwise, the construction of the gene deletion strain had failed. Thus, in the gene deletion strain, the 121 bp fragment could not be amplified (lane 4), while the spc gene could be amplified (lane 2). The gene of the wild-type strain did not change, thus, in the wild-type strain, the 121 bp fragment could be amplified (lane 3), but the spc gene could not be amplified (lane 1). (A) PCR amplification of the spc gene and the deleted part of the sprA gene. Lane M: DL2000 DNA Marker. Lane 1: No fragment was amplified from wild-type strain RA-YM using primer pairs spc-F1/spc-F2 for the spc gene; Lane 2: The spc gene (1086 bp) was amplified from mutant strain ΔsprA using primer pairs spc-F1/spc-F2; Lane 3: The deleted fragment (121 bp) was amplified from wild-type strain RA-YM using primer pairs sprA-N1 and sprA-N2; Lane 4: No fragment was amplified from mutant ΔsprA using primer pair sprA-N1 and sprA-N2 for deleted fragment. (B) PCR amplification of complemented strain CΔsprA. Lane M: DL2000 DNA Marker. Lane 1: The deleted fragment (121 bp) was amplified from complemented strain CΔsprA using primer pair sprA-N1 and sprA-N2; Lane 2: The deleted fragment (121 bp) was amplified from wild-type strain RA-YM using primer pair sprA-N1 and sprA-N2; Lane 3: The Amp gene (1019 bp) was amplified from complemented strain CΔsprA using primer pairsbla-F1/bla-F2; Lane 4: No fragment was amplified from wild-type strain RA-YM using primer pairs bla-F1/bla-F2 for Amp gene. (C) Growth curves. The mutant strain ΔsprA, wild-type strain RA-YM and the complemented strain CΔsprA were grown on TSB medium, and growth of each strain was monitored by measuring the OD600 values. (D) Liquefied gelatin following cultivation of RA-YM, ΔsprA, and CΔsprA on a nutrient gelatin plate.
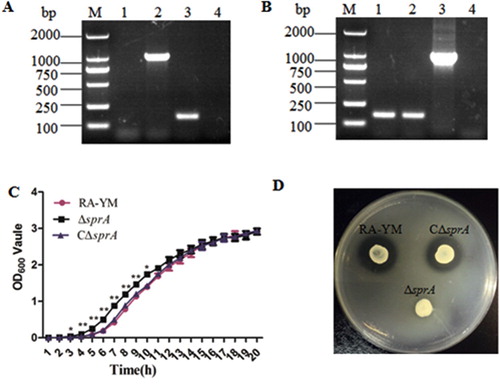
Figure 2. (A) Serum survival of wild-type strain RA-YM, mutant strain ΔsprA, and complemented strain CΔsprA. Effect of sprA mutation on resistance of RA to duck serum. Strains were treated with 12.5% normal duck serum for 30 min at 37°C and plated for survival analysis. Data were analyzed with Student’s t-test (*P < 0.05, **P < 0.01). (B) SDS-PAGE of outer membrane extracts of RA-YM and ΔsprA. M: Protein marker; Lane 1: The outer membrane extract of RA-YM; Lane 2: The outer membrane extract of ΔsprA.
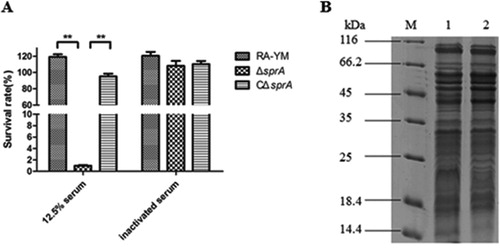
Table 2. Up-regulated outer membrane proteins in mutant strain ΔsprA.
Table 3. Down-regulated outer membrane proteins in mutant strain ΔsprA.
Figure 3. (A) GO annotation for differentially expressed OMPs of up-regulated proteins; (B) Molecular function of up-regulated proteins; (C) GO annotation of down-regulated proteins; (D) Molecular function of down-regulated proteins.
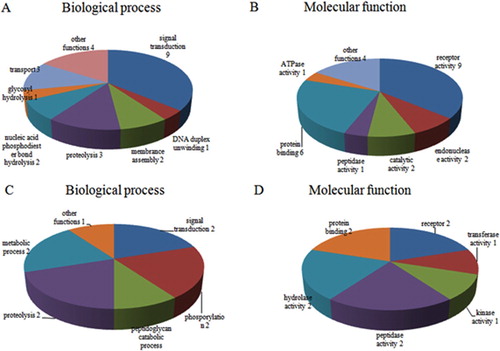
Table 4. CTD proteins identified in RA-YM outer membrance by TMT analysis.
Figure 4. Bacterial loads of tissues. (A) Tissue samples were collected at 24 h post-challenge. (B) Tissue samples were collected at 48 h post-challenge. Data were presented as mean ± standard deviations from five infected ducks. Student’s t-test was used to analyse the data (*P < 0.05, **P < 0.01).
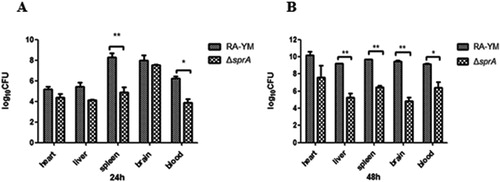
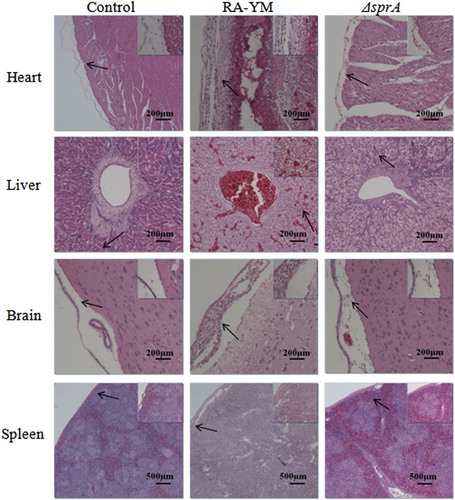
Figure 5. Photomicrograph of histopathological study on tissues from ducklings infected with wild-type strain RA-YM or mutant strain ΔsprA at 24 h post-challenge (HE, × 400). The arrows in the figure indicate the normal (control group) and lesion (RA-YM or ΔsprA infected group) regions of the organs at 24 h post-challenge. Compared with the control group (the left column), there were obvious histopathological changes in the wild-type RA-YM infected group (the middle column): in heart, the visceral pericardium became thickened due to fibrin exudation and inflammatory cell infiltration; in liver, severe fatty degeneration was found, and white pulp lymphocytes hyperplasia occurred in the spleen; no obvious pathological change was found in brain tissue. The histopathological change in the mutant strain ΔsprA infected group (the right column) was slight, thus the tissues from this group looked similar to those of the control group.
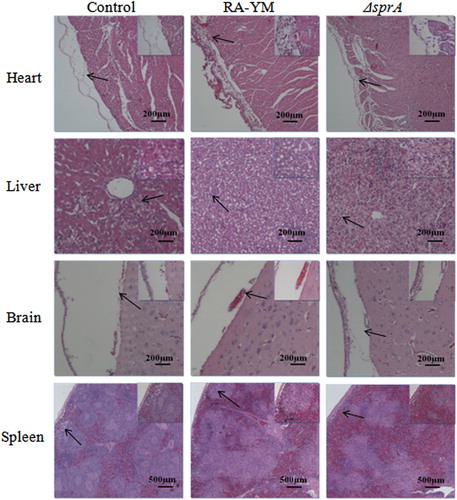
Figure 6. Photomicrograph of histopathological study on tissues from ducklings infected with wild-type strain RA-YM or mutant strain ΔsprA at 48 h post-challenge (HE × 400). The arrows in the figure indicate the normal (control group) and lesion (RA-YM or ΔsprA infected group) regions of the organs at 48 h post-challenge. Compared with the control group (the left column), there were obvious histopathological changes in the wild-type RA-YM infected group (the middle column): in heart, epicardium became thickened due to fibrin exudation and interstitial inflammatory cell infiltration; the spaces between myocardial fibres were significantly widened due to oedema; in liver, moderate to marked congestion was found in central vein and hepatic sinus, and fatty degeneration of the hepatic cells was more severe; in brain, fibrin exudation and inflammatory cell infiltration also occurred in cerebral arachnoid cavity; in spleen, white pulp lymphocyte hyperplasia occurred, and boundaries between white and red pulp became blurred, while germinal centres disappeared. The histopathological change in the mutant strain ΔsprA infected group (the right column) was slight, thus the tissues from this group looked similar to those of the control group.