Figures & data
Table 1. List of primers used to amplify full-length IBDV viral protein 1 (VP1) and VP2 sequences.
Figure 1. Representative histological lesions in bursa sections of chickens infected with Korean antigenic variant infectious bursal disease virus (avIBDV) strains isolated in this study. (a, b) Bursa of 19D38-infected chickens showed mild lymphoid reduction and congestion. (c, d) Bursa follicles of chickens infected with 19D69 were infiltrated with mononuclear inflammatory cells, and showed lymphoid depletion, loss of follicular epithelium, cysts, and connective tissue formation. (e, f) Severe bursal lesions caused by 19D75 including follicle destruction, massive lymphocyte depletion, and granulocyte degeneration. Tissues were subjected to H&E staining, and images were captured at 40× magnification (a, c, e) or 100× magnification (b, d, f).
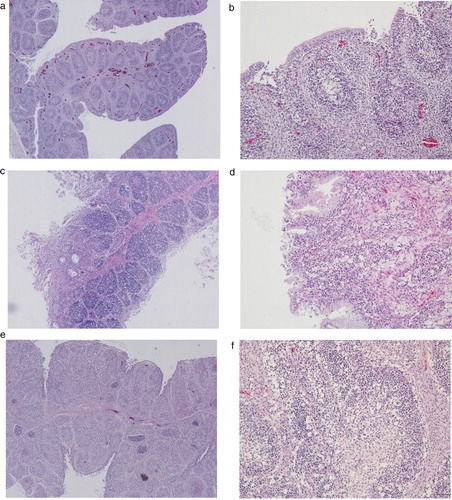
Table 2. Summary of Korean variant strains of IBDV identified in 2019 via passive surveillance of 2–9-week-old chicken carcasses.
Figure 2. Phylogenetic analysis of the nucleotide sequences of viral protein 2 (VP2) and VP1. Phylogenetic trees of hypervariable region of VP2 (a) and partial VP1 (b) generated by the neighbor-joining method using MEGA X with 1000 bootstrap replication. Korean IBDV strains identified in this study are indicated in bold text.
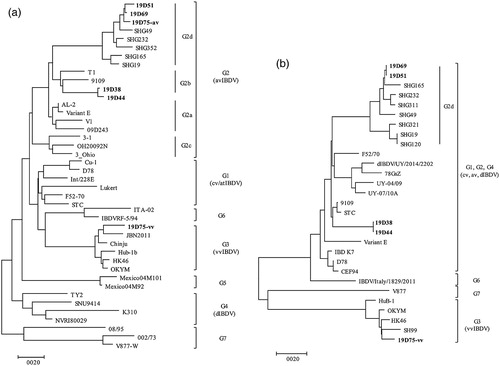
Table 3. Key amino acid residues in the hypervariable region of VP2 showing phylogenic characteristics of Korean antigenic variant IBDVs and reference strains.
