Figures & data
Figure 1. Summary of clinical cases (n = 146) of fowl adenovirus (FAdV) in Korea between 2013 and 2019. (a) Geographical distribution of clinical cases of FAdV. (b) Clinical cases of FAdV associated with different types of chickens. (c) Number and proportion of clinical cases per FAdV serotype. (d) Number and proportion of co-infection cases of FAdVs associated with other pathogens. Number of bacterial co-infection cases: E.coli: 27, Staphylococcus spp: 4, Salmonella spp: 4, Mycoplasma synoviae: 1.
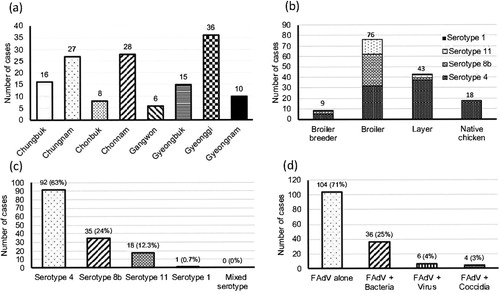
Figure 2. The annual proportion of clinical cases for each FAdV serotype from 2013 to 2019. Absolute numbers of cases are given in parentheses.
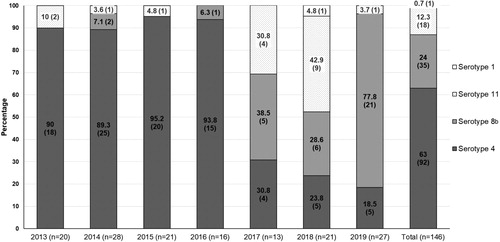
Figure 3. The age of chickens at the time when FAdV infection was confirmed. (a) FAdV-4, (b) FAdV-8b, (c) FAdV-11, (d) FAdV-1. Horizontal lines present the average age of infection.
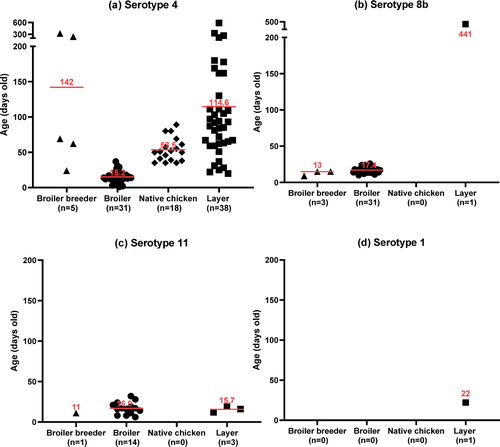
Figure 4. Post mortem and histological examinations of FAdV cases in South Korea. (a), (b), FAdV-8b: (a) Multifocal necrosis in the pancreas; (b) Large necrotic area associated with intranuclear inclusion bodies (INIB, arrow). (c), (d) FAdV-1: (c) Focal erosion and haemorrhage in the ventriculus; (d) Lymphocyte infiltration and INIB in the mucosal area (arrow). (e), (f) IBH: (e) Enlarged liver with multiple petechial haemorrhages; (f) Focal necrosis of hepatocytes associated with basophilic INIB (arrow).
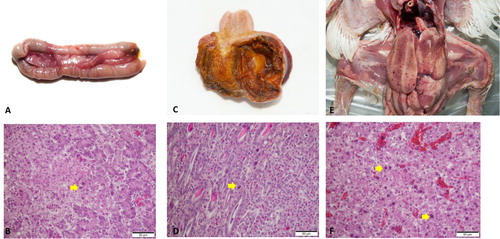
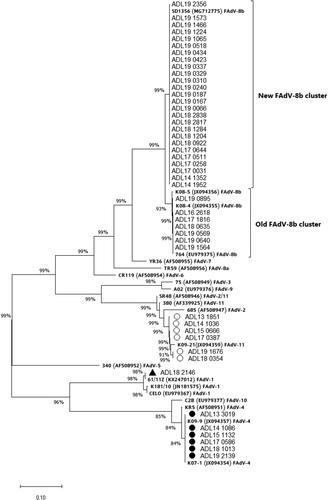
Figure 5. Phylogenetic analysis of FAdV isolates based on the nucleotide sequences of the hexon loop-1 gene. The tree was constructed by the maximum likelihood (ML) method with Mega version 10. The strains sequenced for this study are designated by names starting with ADL. Reference strains are in bold text with accession numbers in parentheses. For the FAdV-4 and 11 groups, a strain isolated from each year (2013–2019) was selected and listed in the tree as a representative. Solid circles indicate representative strains for the isolated FAdV-4 serotypes. Representative strains for the isolated FAdV-11 serotypes are indicated by open circles. The only FAdV-1 strain (ADL18 2146) from this study is indicated by a solid triangle. The old and the newly formed clusters of FAdV-8b are shown on the right. Sequence data of the isolated FAdV strains were submitted to GenBank under the accession numbers MN737046-MN737093. SR48 (AF508949) is officially designated as FAdV-2; however, recent publications suggest a reclassification of this strain into FAdV-11 (Steer et al., Citation2011; Marek et al., Citation2016; Feichtner et al., Citation2018; Schachner et al., Citation2019).