Figures & data
Table 1. Case summary and detection of H9N2 strains from field cases.
Figure 1. Individual egg production rate changes of farms affected by the new H9N2 field isolates. Time points of field sera and necropsy sample collection are indicated by arrows.
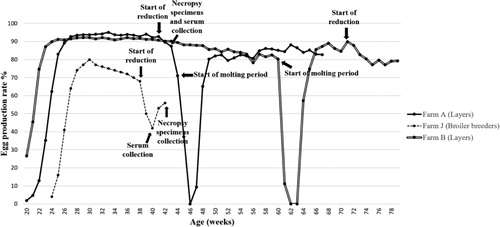
Figure 2. Lesions from post-mortem examinations of new H9N2 Y280-like LPAI cases in South Korea. (a) Fatty liver haemorrhagic syndrome (FLHS). (b) Necrotic and vacuolated hepatocytes. (c) Primary bronchi with caseous plugs (arrow). (d) Lymphocyte infiltration and cell necrosis in the mucosa with vacuoles containing cell debris in the trachea of an infected broiler. Loss of cilia was observed. The tracheal sample was positive for infectious bronchitis (IB) virus and H9N2 avian influenza (AI) virus by RT-PCR.
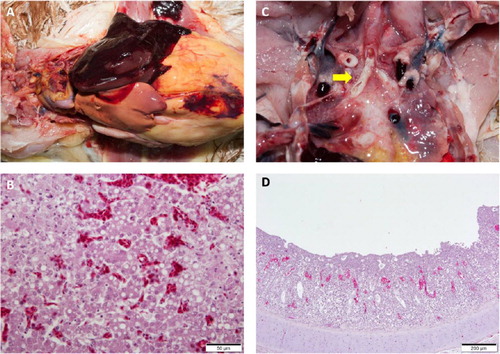
Figure 3. Phylogenetic analysis of the H9N2 Y280-like field isolates based on the nucleotide sequences of the haemagglutinin (HA) gene. The tree was constructed by the maximum likelihood (ML) method with Mega version 10. Black dots indicate H9N2 field isolates from this study. The A/chicken/Shandong/1844/2019 strain is indicated by a white dot. The representative strains for the Y280-like lineage, the Y439/Korean lineage and the G1 lineage are indicated by a black triangle, diamond and square, respectively. The strains from the Y439/Korean lineage and Y280-like lineage are shown on the right. Sequence data of the new H9N2 field isolates were submitted to GenBank under the accession numbers MW570860-MW570867.
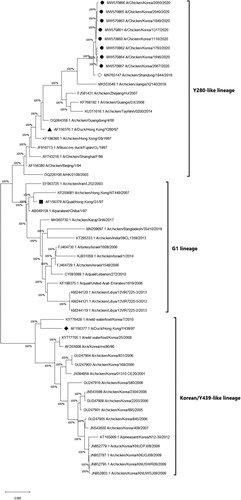
Figure 4. Phylogenetic analysis of the H9N2 Y280-like field isolates based on the nucleotide sequences of the neuraminidase (NA) gene. The tree was constructed by the maximum likelihood (ML) method with Mega version 10. Black dots indicate H9N2 field isolates from this study. The A/chicken/Shandong/1844/2019 strain is indicated by a white dot. The representative strains for the Y280-like lineage, Y439/Korean lineage and G1 lineage are indicated by a black triangle, diamond and a reversed white triangle, respectively. The NA gene of the A/chicken/Korea/580/2006 strain (black square) is known to have the highest sequence identity with the Korean live bird market H3N2-like lineage, not with H9N2 strains (Lee et al., Citation2012). Sequence data of the new H9N2 field isolates were submitted to GenBank under the accession numbers MW570852-MW570859.
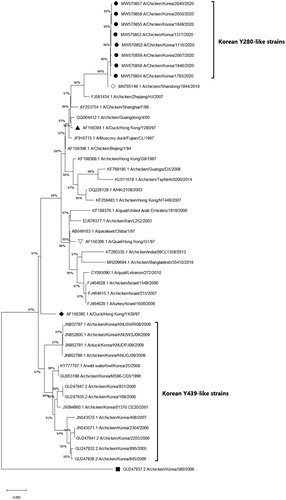
Table 2. Cleavage site and receptor-binding pocket of H9N2 field isolates and reference strains.
Table 3. Potential N-glycosylation sites (PGS) of the H9N2 field isolates and reference strains.
Table 4. Characteristics of field sera collected from infected farms.
