Figures & data
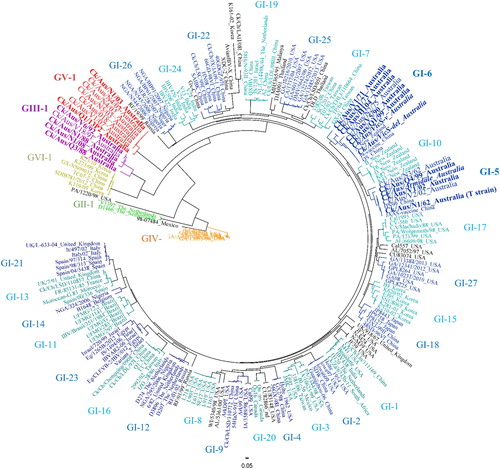
Figure 1. Timeline showing the emergence of the different infectious bronchitis virus lineages in Australia and the introduction of attenuated vaccines using selected strains belonging to those lineages.
Table 1. Infectious bronchitis virus strains isolated in Australia.
Figure 2. Comparison of two phylogenetic trees constructed with the same multiple alignment of the complete genome sequences of Australian strains of IBV. The multiple alignment was performed using MAFFT, and the phylogenetic trees using PHYML (maximum likelihood) and MrBayes (Bayesian) in Geneious v10.2.6. Two phylogenetic trees were included using two different algorithms to increase reliability of the results. Originally, the complete genome nucleotide sequences of 19 Australian strains were included in this tree. However, some strains were removed to increase clarity, as they were highly similar or identical to other strains. The strains not included were H104 (100% similarity to Q1/73), Q1/99, VicS-del and VicS-v (99.9%, 99.7% and 99.9% of similarity with V5/90). The grey squares depict the different genotypes GI, GIII and GV.
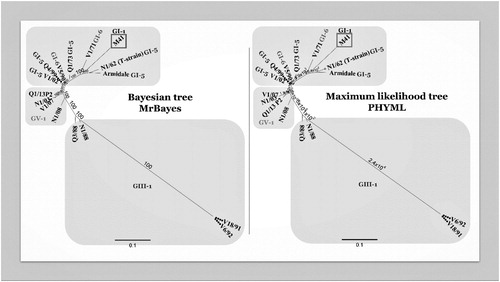
Figure 3. Radial phylogenetic tree based on the complete nucleotide sequence of the IBV surface protein S1. Due to its complexity, GI genotype was subdivided in 27 lineages. Different colours represent different genotypes (light and dark blue are considered the same colour, both belonging to GI genotype). The names of unclassified strains are in black. The names of the Australian strains are magnified. Only Australian strains with a complete nucleotide sequence of the S1 gene were included. The vaccine strains A3 (Armidale or N1/62) and VicS (represented by the two subpopulations VicS-v and VicS-del) are rendered in italics.