Figures & data
Figure 1. Human Cytochrome P450 1A1 in complex with alpha-naphthoflavone. Exon 2 (1-275) rainbow blue extends from the N-terminus to G-H Loop; Exon 3 (276-317) G-H Loop to Helix I; Exon 4 (318-347) Helix I to Helix J; Exon 5 (348- 389) Helix J to β1-4 sheet, Exon 6 (390-418) Brain-specific β1-4 sheet to K’-K” Loop; and Exon 7 (419-512) K’-K” to C-terminus red. PDB ID: 4I8V (Walsh et al. Citation2013; Annalora et al. Citation2017; Rose et al. Citation2018).
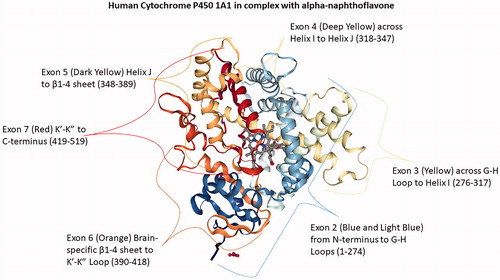
Figure 2. The expression of human cytochrome P450 enzymes involved in the metabolism of xenobiotics in the selected brain regions shown in coronal, sagittal, and horizontal brain sections.
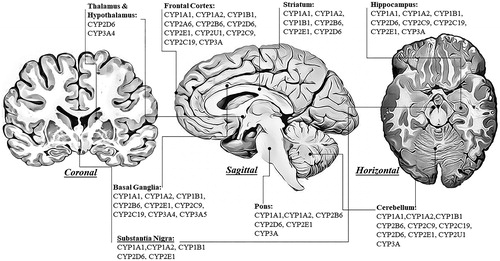
Figure 3. An overview of human CYP1A cluster, located on chromosome 15q22-qter with CYP1A1 reading toward the telomere and CYP1A2 reading toward the centromere (Corchero et al. Citation2001). The distance between the start of transcription of CYP1A1 and CYP1A2 is 23.3 kb. There are 13 XRE (xenobiotic‐responsive element) at the core DNA sequences that may participate in CYP1A induction following activation of the Ah receptor (adapted from Galijatovic et al. Citation2004; Ye et al. Citation2019).
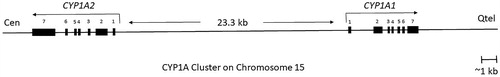
Figure 4. An overview of the CYP2A-T gene cluster on human chromosome 19. Arrows and blocks represent the position, relative length and direction of transcription for each locus (adapted from Hoffman et al. Citation2001), ‘‘P’’ at the end of a gene name indicates a pseudogene.

Figure 5. An outline of the CYP2D gene cluster on human chromosome 22. The cluster contains the CYP2D6 functional gene and two or more highly homologous pseudogenes CYP2D8P and CYP2D7, the gene CYP2D6 is highly polymorphic, and there is substantial variation in allele occurrences between people residing in different geographical location. Arrows and blocks represent the position, relative length, and direction of transcription for each locus, vertical line separate single copy from duplicated type. 5.5% of the Europeans carry more than two active CYP2D6 gene copies and are ultrarapid metabolisers (UMs), whereas the mutation is essentially absent in Asia (adapted from Koch Citation2004; Ingelman-Sundberg Citation2005).
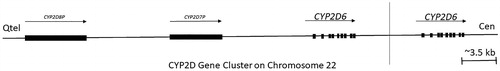
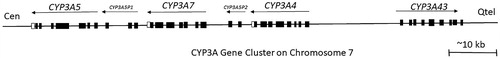
Figure 6. The arrangement of the CYP3A gene cluster with pseudogenes at the CYP3A locus. Abbreviations: Cen - centromere; Qtel - telomere. Exons for genes and pseudogenes are shown as boxes; arrows indicate transcriptional orientation (adapted from Chen et al. Citation2009).