Figures & data
Figure 1. The majority subcellular locations of CYPs and UGTs in brain cell. The expression of CYPs was detected on the plasma membrane, but mainly in the cytoplasmic side of ER (Zhang et al. Citation2016) and the inner membrane of the mitochondria (Walther et al. Citation1986). However, the substrate binding sites are deeply buried in the phospholipid bilayer membranes (Šrejber et al. Citation2018). UGTs are predominantly located on the luminal side of the ER (Ouzzine et al. Citation2014).
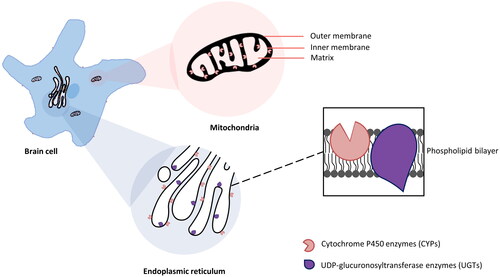
Table 1. Distribution of major CYP family DMEs in different regions and cell types in human, rat, and mouse brains rat, mouse and human CYPs that are present in several distinct brain localizations.
Table 2. Distribution of major UGT family DMEs in different regions and cell types in human, rat, and mouse brains rat, mouse and human UGT DMEs that are present in several distinct brain localizations.
Table 3. Major CYP family DME genes in human, rat, and mouse brains.
Table 4. Major UGT family DME genes in human, rat, and mouse brains.
Table 5. Comparison of ex vivo and in vivo enzyme activity measurement methods.
Table 6. DMEs in human brains and corresponding CNS-active drugs as substrates.
