Figures & data
Table 1. Wheat genotypes used to differentiate races of Puccinia striiformis f. sp. tritici in the United States
Fig. 1 Distributions and its relative severity of wheat stripe rust, caused by Puccinia striiformis f. sp. tritici, in the United States from 2000 to 2007.
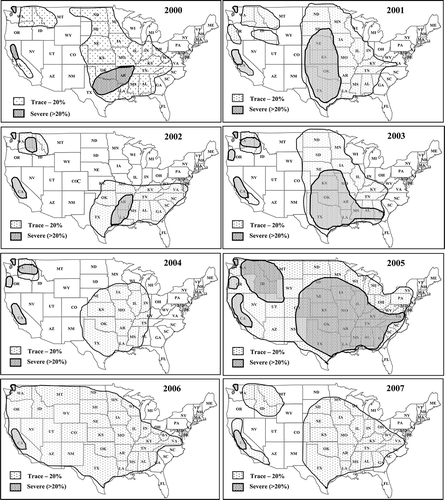
Fig. 2 Epidemiological regions of stripe rust of wheat, caused by Puccinia striiformis f. sp. tritici, in the United States and Canada (see Line & Qayoum (Citation1992) for details). The previous region 7 (separated from other regions in the west by the line along the Rocky Mountains) is divided into regions 7 to 12. Region 1 (R1) = eastern Washington, northeastern Oregon, and northern Idaho; R2 = western Montana and southern Alberta, Canada; R3 = southern Idaho, southeastern Oregon, northern Nevada, northern Utah, western Wyoming, and western Colorado; R4 = western Oregon and northern California; R5 = northwestern Washington and southwestern British Columbia, Canada; R6 = central and southern California, Arizona, and western New Mexico; and R7 = Texas, Louisiana, Arkansas, Oklahoma, and eastern New Mexico; R8 = Kansas, Nebraska, and eastern Colorado; R9 = South Dakota, North Dakota, Minnesota, eastern Montana, and southern Manitoba and southern Saskatchewan, Canada; R10 = Mississippi, Alabama, Florida, Georgia, South Dakota, North Dakota, Tennessee, and Kentucky; R11 = Missouri, Illinois, Indiana, Iowa, Wisconsin, Michigan, and Ontario, Canada; and R12 = Virginia, West Virginia, Ohio, Maryland, Pennsylvania and New York.
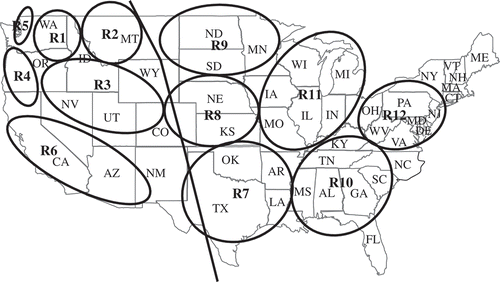
Table 2. Numbers of viable isolates, races, and first detected races and the most predominant races of Puccinia striiformis f. sp. tritici in the United States from 2000 to 2007
Table 3. Virulences, the first year detected, and frequencies and distribution of races of Puccinia striiformis f. sp. tritici (PST) in the United States from 2000 to 2007
Fig. 3 Top five predominant races of Puccinia striiformis f. sp. tritici and their frequencies in the United States from 2000 to 2007.
Table 4. Frequencies of virulence factors in Puccinia striiformis f. sp. tritici from 2000 to 2007
Fig. 4 Frequency changes of virulence factors to the 20 wheat genotypes that were used to differentiate races of Puccinia striiformis f. sp. tritici in the United States from 2000 to 2007, showing a, virulence factors (1, 2, 6, and 12) with relatively stable frequencies; b, those (4, 5, 9, 13, 14, and 15) with frequencies changing from 0–10% to 4–80%; c, those (10, 17, 19 and 20) with frequencies changing from 10–30% to 80–100%; and d, those (3, 8, 11, 16, and 18) from 30–65% to 80–100%. The numbers in the figure represent the virulence factors corresponding to wheat differential number listed in ; 1 = ‘Lemhi’, 2 = ‘Chinese 166’, 3 = ‘Heines VII’, 4 = ‘Moro’, 5 = ‘Paha’, 6 = ‘Druchamp’, 7 = AVS/6*Yr5, 8 = ‘Produra’, 9 = ‘Yamhil'l, 10 = ‘Stephens’, 11 = ‘Lee’, 12 = ‘Fielder’, 13 = ‘Tyee’, 14 = ‘Tres’, 15 = ‘Hyak’, 16 = ‘Express’, 17 = AVS/6*Yr8, 18 = AVS/6*Yr9, 19 = ‘Clement’ and 20 = ‘Compair’.
Fig. 5 Dendrogram of 115 races of Puccinia striiformis f. sp. tritici that were detected in the United States from 2000 to 2007 that have been arranged based on their virulence and avirulence to the set of 20 wheat differential genotypes, using the unweighted pair group arithmetic mean (UPGMA) program of NTSYS-pc (version 2.2.1; Rohlf, Citation1992).
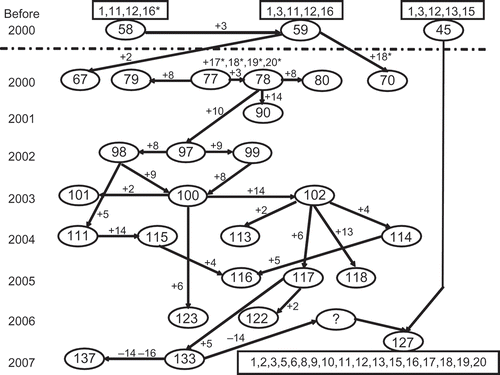
Fig. 6 Postulated evolutionary relationships of selected races of Puccinia striiformis f. sp. tritici to demonstrate the major virulence changes in the US population of the pathogen from 2000 to 2007. The numbers in oval cycles represent PST races (see ). The numbers in a rectangle represent the virulence formula for the race (see ). ‘+’ indicates addition of one or more virulence factors and ‘-‘ indicates loss of one or more virulence factors to the wheat differentials. Wheat differential numbers () are: 1 = ‘Lemhi’, 2 = ‘Chinese 166’, 3 = ‘Heines VII’, 4 = ‘Moro’, 5 = ‘Paha’, 6 = ‘Druchamp’, 7 = AVS/6*Yr5, 8 = ‘Produra’, 9 = ‘Yamhill’, 10 = ‘Stephens’, 11 = ‘Lee’, 12 = ‘Fielder’, 13 = ‘Tyee’, 14 = ‘Tres’, 15 = ‘Hyak’, 16 = ‘Express’, 17 = AVS/6*Yr8, 18 = AVS/6*Yr9, 19 = ‘Clement’ and 20 = ‘Compair’.