Figures & data
Table 1. Isolates of Pyrenophora tritici-repentis analyzed in this study, including isolate name, race designation and geographic origin
Table 2. Simple sequence repeat loci identified in the genome of Pyrenophora tritici-repentis (Broad Institute), including motif length and copy number
Table 3. Simple sequence repeat loci utilized to assess genetic diversity and relatedness in a global collection of isolates of Pyrenophora tritici-repentis
Table 4. Simple sequence repeat (SSR) polymorphic loci and genetic diversity
Fig 1. Unrooted UPGMA tree based on Nei's genetic distance between populations of Pyrenophora tritici-repentis from Syria (SYR), Algeria (ALG), Canada (CAN) and Azerbaijan (AZ). The scale bar represents 1 substitution per site. UPGMA = unweighted pair group method using arithmetic averages.
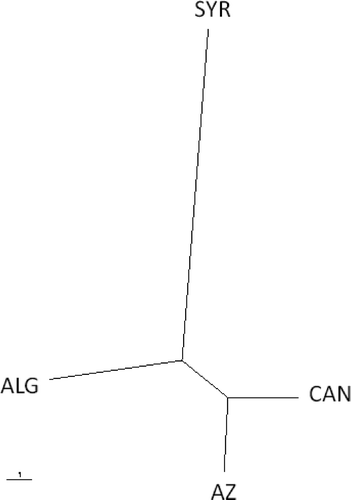
Fig 2. Unrooted UPGMA tree based on Nei's genetic distance between races 1, 2, 3, 5, 7 and 8 of Pyrenophora tritici-repentis. Races 4 and 6 were excluded from the analysis because only a very limited number of isolates from these races were available. The scale bar represents 1 substitution per site. UPGMA = unweighted pair group method using arithmetic averages.
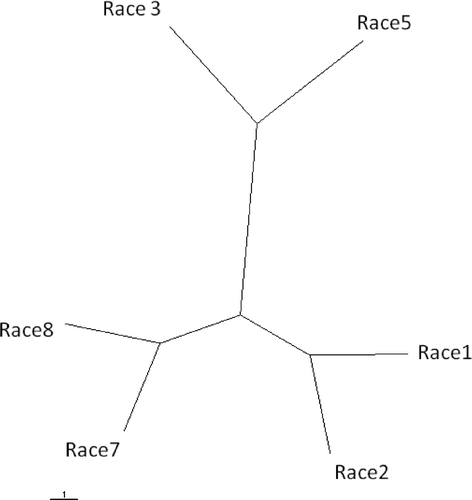
Table 5. Analysis of molecular variance between populations of Pyrenophora tritici-repentis originating from different countries
Table 6. Analysis of molecular variance in different races of Pyrenophora tritici-repentis
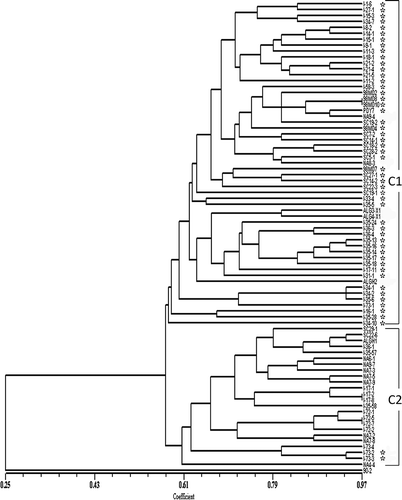
Fig 3. Dendrogram obtained by UPGMA analysis showing the genetic similarity of a global collection of isolates of Pyrenophora tritici-repentis. The vertical lines define the two main clusters. Asterisks denote Ptr ToxA-producing isolates of the fungus. UPGMA = unweighted pair group method using arithmetic averages.