Figures & data
Fig. 1. PCR detection of phytoplasmas using the universal primer pair (P1/P7). Templates consisted of DNA extracted from periwinkle plants infected with the following phytoplasma reference strains: apple proliferation (AP); European stone fruit yellows (ESFY) and pear decline (PD) or from symptomless plum (H) and symptomatic plums (1–5). W, water control; M, marker.
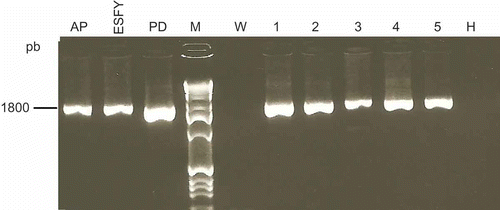
Fig. 2. Group-specific PCR amplifications of phytoplasmas using the 16SrX specific primer pair (fO1/rO1). Templates consisted of DNA extracted from periwinkle plants infected with the following phytoplasma reference strains: apple proliferation (AP); European stone fruit yellows (ESFY) and pear decline (PD) or from symptomless plum (H) and symptomatic plums (1–5). W, water control; M, marker.
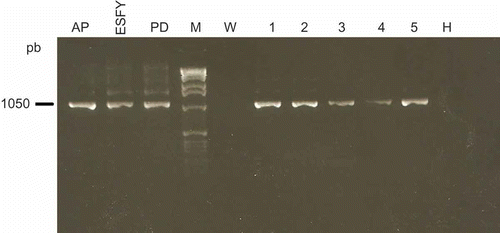
Fig. 3. SspI restriction profiles of phytoplasma ribosomal DNA amplified using the universal primer pair P1/P7. Template DNA was from periwinkle plants infected with phytoplasma reference strains: pear decline (PD); European stone fruit yellows (ESFY) and apple proliferation (AP) or from symptomatic plums (1–5). M, marker.
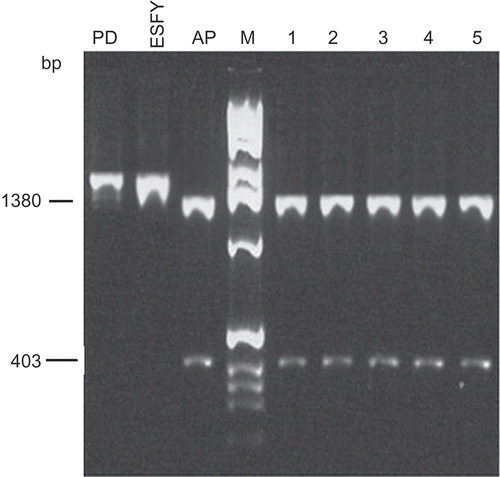
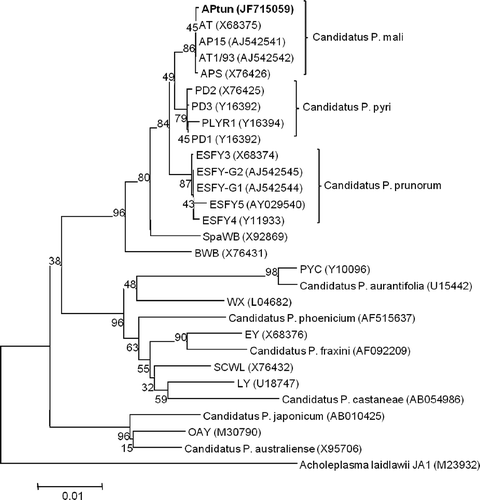
Fig. 4. Phylogenetic tree constructed by using the neighbour-joining method with 16S rDNA sequences from strain (APtun) detected in plum decline in Tunisia (bold), strains of ‘Candidatus P. pyri’ (PD1–PD3 and strain PYLR1 from PYLR phytoplasma), ‘Candidatus P. mali’ (AP15, APS, AT and AT1/93) and ‘Candidatus P. prunorum’ (ESFY-G1, ESFY-G2, ESFY3–ESFY5) and 14 reference phytoplasmas. Acholeplasma laidlawii was used as the outgroup.