Figures & data
Table 1. Bacteria used in this study and specificity of detection of Pseudomonas syringae pathovars by TaqMan real-time and conventional polymerase chain reactions
Fig. 1. Sensitivity of detection of purified DNA of Pseudomonas syringae pv. syringae 99_5 by TaqMan real-time PCR assay targeting the cyoA gene. a, Fluorescence kinetics and b, standard curve showing log of DNA dilutions and the corresponding cycle threshold (CT). The regression equation and coefficient of determination are indicated. Each dot represents data of triplicate TaqMan real-time PCR amplifications.
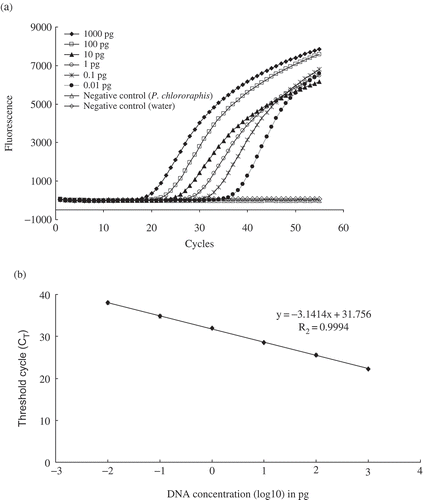
Fig. 2. Sensitivity of detection of serially diluted cultures of Pseudomonas syringe 99_5 by TaqMan real time PCR assay. One microlitre of the bacterial suspension was assayed. Mean viable plate count, colony forming units (CFU) mL−1: sample 1, 4.5 × 108; sample 2, 4.5 × 107; sample 3, 4.5 × 106; sample 4, 4.5 × 105; sample 5, 4.5 × 104; sample 6, 4.5 × 103; sample 7, 4.5 × 106 Pseudomonas chlororaphis PCL1391; sample 8, negative control (autoclaved milliQ water).
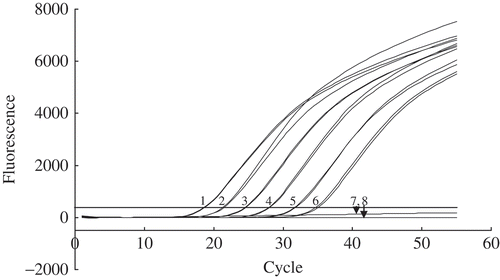
Fig. 3. a, Determination of real-time PCR inhibition in direct detection of Pseudomonas syringae in leaf crude extracts of tomato. Ten-fold serial dilutions of tomato leaf crude extracts spiked with 1 ng of purified bacteriophage lambda DNA were analyzed using TaqMan real-time PCR as described previously (Tambong et al., Citation2008). Threshold cycle (CT) values are inversely proportional to the amount of DNA in a sample. Higher CT value for undiluted tomato leaf crude extract is an indication of real-time PCR inhibition. Each bar is a mean of three replicates. b, Detection by TaqMan real-time PCR of Pseudomonas syringae pv. tomato DC3000 of (a) Purified genomic DNA or (b) cells on tomato leaves inoculated under growth chamber condition; (c) non-inoculated tomato leaves and (d) Negative control water. No DNA extraction performed. Six plants were analyzed using TaqMan real-time PCR in triplicates. Leaf lesions were processed as described in Materials and methods. The presence of Pseudomonas syringae on infected tomato leaves was confirmed by isolation on agar medium and by conventional PCR amplifications of specific DNA fragment of cytochrome o ubiquinol oxidase. The experiment was repeated once with similar results.
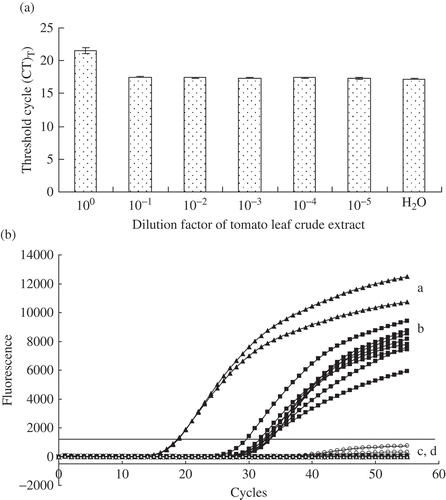
Fig. 4. Partial alignments of cytochrome o ubiquinol oxidase subunit II gene sequences showing nucleotide mismatch(es) in TaqMan probe (a) and forward primer (b). Primer and probe were designed from Pseudomonas syringae pv. syringae B728a genome as the reference. Nucleotide(s) in a box depict(s) a mismatch relative to the reference genome and primer or probe. Sequences of strains were generated as reported in the Materials and methods. Note nucleotide mismatch for the probe is almost at the centre; and for the forward primer, there is a crucial mismatch at the 3'-end. Asterik (*) strain tentatively identified as P. syringae pv. syringae (D. Cuppels, pers. comm.).

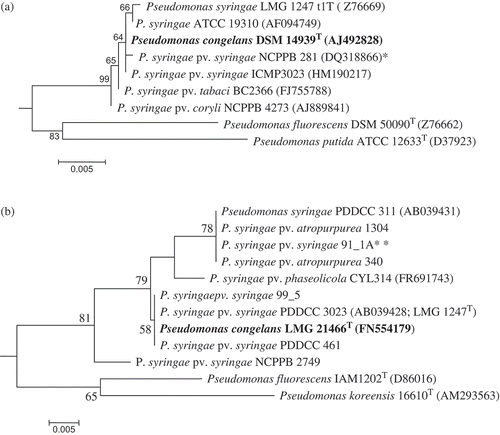
Fig. 5. a, 16S rRNA and b, gyrase B maximum likelihood (ML) phylogeny reconstructions revealed a close relationship between Pseudomonas syringae and Pseudomonas congelans. 16S rRNA (1290 positions) phylogeny was implemented using the General Time Reversible model while gyrase B DNA sequences were translated to amino acids (183 positions) and phylogeny inferred using the Poisson correction model. Both phylogenies were inferred using MEGA4 (Tamura et al., Citation2007) with bootstrap values (1000 replicates) shown next to the branch nodes. Trees were rooted with Escherichia coli (X80725) for 16S rRNA and E. coli E1140 (HQ660616) for gyrase B. Asterik(s) indicates the neopathotype strain (*) or a strain tentatively identified as pathovar syringae (**).