Figures & data
Table 1. The origin of country, host and GenBank accession numbers of the MP and CP genes of PNRSV isolates used for phylogenetic analyses
Fig. 1. RT-PCR detection of Prunus necrotic ringspot virus (PNRSV) isolates from Canada. (a), Amplified PCR products showing the near-full-length genomic RNA3 segment with primers RNA-R and RNA3-F. (b), Products with primers RNA-R and RNA3-F1. M, molecular weight markers; 1, water control; 2, negative control; 3 through 19, Chr1 through Chr18 (except Chr3); 20 through 31, Pch1 through Pch13 (except Pch12). Isolates Chr3 and Pch12 were shown positive in initial RT-PCR tests with primers RNA-R and RNA3-F and were not repeated here. M: 1kb DNA ladder marker.
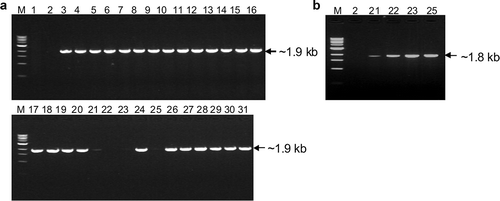
Fig. 2. A pair-wise alignment of movement protein sequences of 10 most divergent isolates from this study and representative isolates from four phylogenetic groups. The same and deleted amino acids are indicated as · and Δ, respectively.
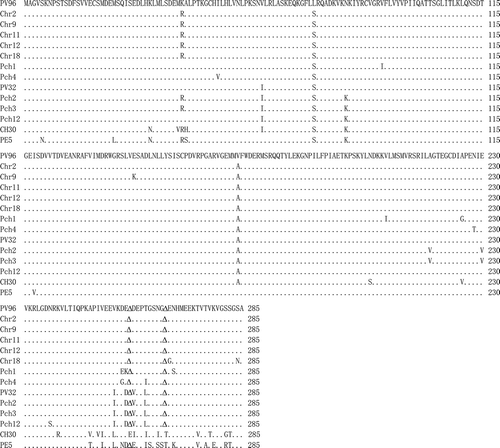
Fig. 3. A pair-wise alignment of coat protein sequences of 10 most divergent isolates from this study and representative isolates from four phylogenetic groups. The same and deleted amino acids are indicated as · and Δ, respectively.

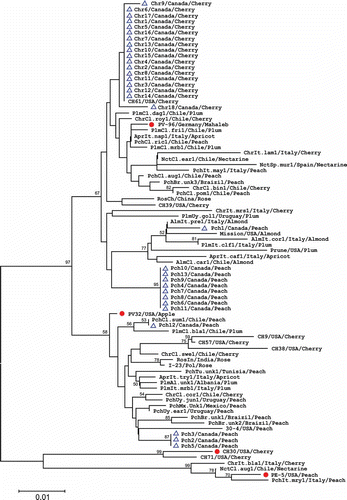
Fig. 4. Phylogenetic tree based on the CP amino acid sequences of 31 Prunus necrotic ringspot virus (PNRSV) isolates from Canada and 53 isolates retrieved from the NCBI database. Four PNRSV isolates representing phylogroups PV96, PV32, CH30 and PE5 are indicated by the symbol •, and all 31 Canadian isolates are marked by ▴. The tree was reconstructed with MEGA 4.1 using the neighbour-joining method. The numbers at the nodes indicate the percentage of 1000 bootstraps occurred in this group. The scale bar represents the number of substitutions per amino acid.
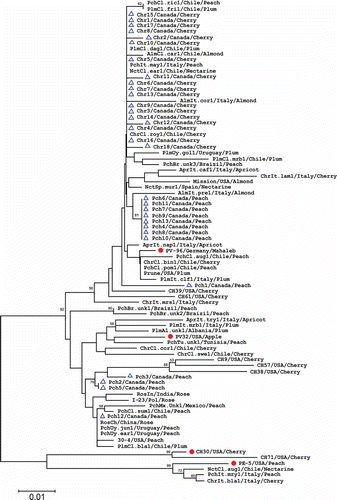
Fig. 5. Phylogenetic tree based on the MP amino acid sequences of 31 Prunus necrotic ringspot virus (PNRSV) isolates from Canada and 53 isolates retrieved from the NCBI database. Four PNRSV isolates representing phylogroups PV96, PV32, CH30 and PE5 are indicated by the symbol •, and all 31 Canadian isolates are marked by ▴. The tree was reconstructed with MEGA 4.1 using the neighbour-joining method. The numbers at the nodes indicate the percentage of 1000 bootstraps occurred in this group. The scale bar represents the number of substitutions per amino acid.