Figures & data
Table 1. Number of actinobacteria isolated from various Algerian rhizospheric soil
Table 2. Fungal plant pathogens used in antagonism assays
Table 3. Morphological and physiological characteristics of the six antagonistic actinobacteria isolates
Fig. 1. Effect of antagonistic actinobacterial isolates on mycelium growth of phytopathogenic fungi. Results are the means of triplicates. Values from bars with the same letter do not differ statistically (LSD). (a) Fusarium culmorum, (b) Microdochium nivale, (c) Fusarium oxysporum, (d) Drechslera teres, (e) Verticillium dahliae, (f) Bipolaris sorokiniana and (g) Botrytis fabae.
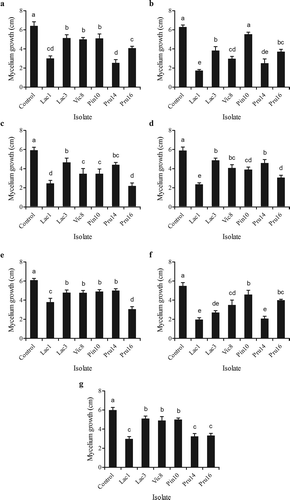
Fig. 2. Spore chain morphology of (a) Streptomyces griseus Lac1, (b) Streptomyces rochei Lac3, (c) Streptomyces anulatus Pru14, (d) Streptomyces champavatii Pru16, (e) Nocardiopsis dassonvillei subsp. dassonvillei Vic8 and (f) Nocardiopsis alba Pin10 observed with scanning electron microscopy. Bars represent 1 μm.
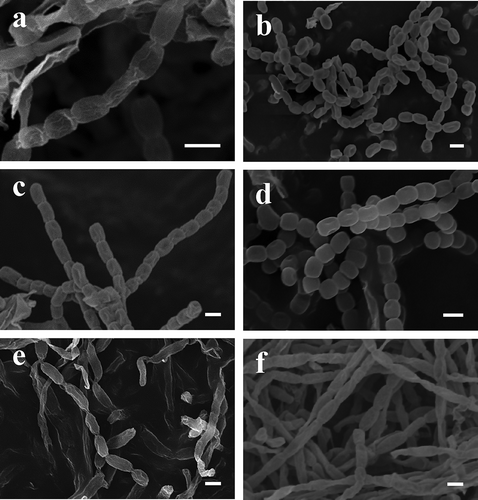
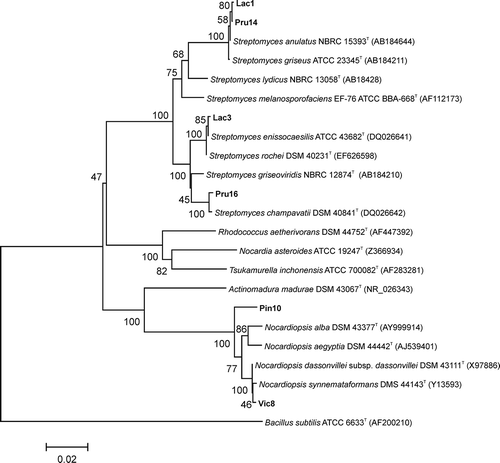
Fig. 3. Phylogenetic tree based on 16S rRNA gene sequences of the six antagonistic strains isolated in this study and their closest relatives. The tree was constructed using the neighbour-joining algorithm. Bootstrap values based on 1000 replicates are shown at the nodes of the tree. The bar indicates a distance of 0.02 substitutions per nucleotide position. Bacillus subtilis was used as an outgroup.