Figures & data
Table 1. Primers used in this study
Table 2. Fungi used in tblastn searches
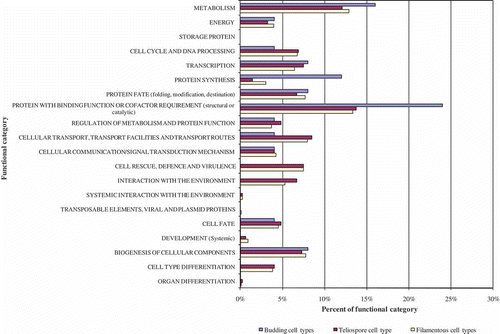
Table 3. EST representation among cDNA libraries and functional associations
Fig. 1. RT-PCR for genes putatively identified as (a) dikaryon specific, (b) constitutively expressed. M: marker; DIK: dikaryon; D132: diploid; f+: filamentous; f-: non-filamentous; TDO: dormant teliospore; FB1, FB2: haploid cells; gDNA: genomic DNA; NTC: no template control.
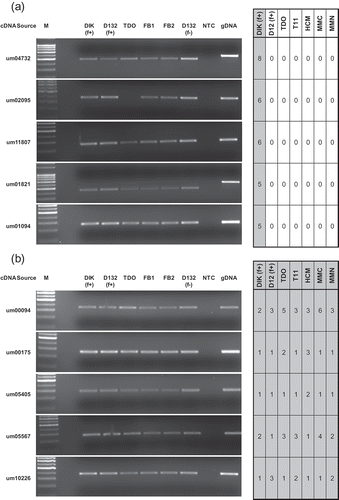
Fig. 2. Controls for RT-PCR analysis. M: marker; DIK: dikaryon; D132: diploid; f+: filamentous; f-: non-filamentous; TDO: dormant teliospore; FB1, FB2: haploid cells; gDNA: genomic DNA; NTC: no template control.
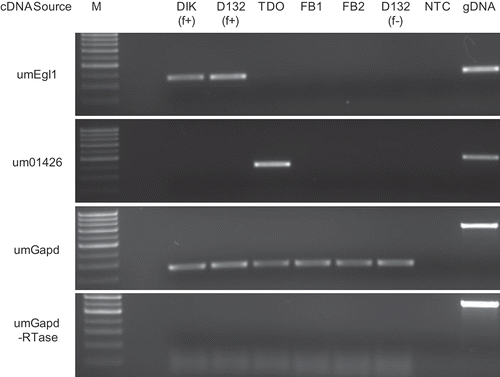
Fig. 4. RT-PCR assessing U. maydis transcript levels in different cell types and during in planta pathogenic development. Genes putatively identified as (a) Ustilago maydis specific, or (b) pathogen specific. (c) The genes ncRNA1 and um12316. (d) RT-PCR controls. M: marker; DIK: dikaryon; D132: diploid; f+: filamentous; TDO: dormant teliospore; FB1, FB2: haploid cells; DPI: days post infection; gDNA: genomic DNA; NTC: no template control.
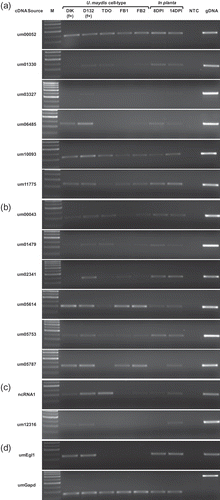
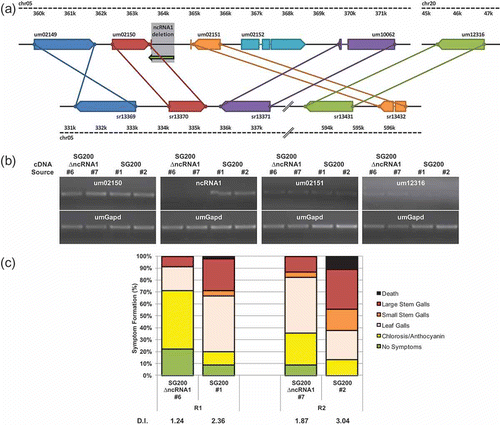
Fig. 5. Deletion of ncRNA1 attenuates pathogenicity. (a) Schematic of the ncRNA1 locus. Regions of U. maydis (top) and S. reilianum (bottom) chromosomes (chr) are represented by dashed black lines. A region of S. reilianum chr05 not shown is represented by diagonal black double-lines. Annotated ORFs are represented by pentagons; putative 3′ UTRs surrounding the ncRNA1 deletion by red and orange vertical lines; intergenic regions by solid black lines; introns by solid coloured lines and the noncoding transcript (ncRNA1) by a green arrow. The region removed from ΔncRNA1 strains is highlighted in grey. Putative orthologues shared between U. maydis and S. reilianum are indicated by solid lines of the same colour. (b) RT-PCR examining the effect of ΔncRNA1 on select transcript levels. RNA was isolated from ΔncRNA1 #6 and ΔncRNA1 #7 with independent SG200 controls (#1 and #2). Transcript-specific primers were used to generate cDNA for um02150, ncRNA1, um02151 and um12316 with an internal umGapd-specific primer to control for relative amounts of RNA. ‘No reverse transcriptase’ and ‘no template’ controls were also performed (data not shown). (c) Pathogenicity of ΔncRNA1 U. maydis strains. Infection with independent ΔncRNA1 strains and their corresponding control (SG200) were performed in parallel. Seven-day-old maize seedlings (n = 45) were infected and the symptoms were scored 14 DPI. Separate infection assays (R1 and R2, respectively) differed slightly in the manifestation of disease symptoms. Bars represent the percentage of infected plants and the disease symptoms are depicted in the legend. The disease index (DI) for each strain is shown below the bar graph.