Figures & data
Table 1. Primers used in the construction and manipulation of F. graminearum transgenic strains and qRT-PCR reactions
Fig. 1 Alignment of FGSG_03881, FGSG_11025, and FS_Tri15 proteins. Amino acid sequences were aligned by ClustalW2. Identical amino acids between all three proteins are highlighted in black, identities between FGSG_03881 and FS_Tri15 are highlighted in light grey, identities between FGSG_11025 and FS_Tri15 highlighted in dark grey, and the zinc fingers are labelled Zn1 and Zn2, respectively.
Fig. 2 The Δfgsg_03881 strain is more virulent at early stages of infection than the wildtype F. graminearum strain. (a) PCR confirmation of the disruption of FGSG_03881. Genomic DNA was extracted from the Δfgsg_03881 (1) and the wildtype (2) strains and amplified with primers specific to hygromycin marker gene (Hyg) and primers specific to FGSG_03881. (b) PCR confirmation of the complement Δfgsg_03381/FGSG_03881 strain. Genomic DNA was extracted from the wildtype (1) and Δfgsg_03381/FGSG_03881 (2) strains and amplified with primers specific to the Geneticin marker gene (Geneticin) and FGSG_03881. The primers used are listed in . (c) A susceptible cultivar of wheat ‘Roblin’ was point inoculated with the wildtype, Δfgsg_03881, and the complement Δfgsg_03381/FGSG_03881 strains and scored for disease symptoms after 7, 10, 14 and 21 dpi. The number of wheat heads inoculated with each strain is indicated by ‘n’. The number of wheat heads with symptoms beyond the inoculation site is represented as percentage infection (%infection). Error bars reflect the standard error and the ‘P values’ were calculated by one-tailed unpaired t-tests comparing the wildtype and test strains. Significant ‘P values’ are bolded to highlight observed infection differences.
Fig. 3 FGSG_03881 expression is dependent on nitrogen limitation and is independent of Tri6. (a) The expression of FGSG_03881, Tri6, and Tri10 is dependent on nitrogen. The wildtype strain was grown in nutrient rich medium (GYEP) for 24 h and transferred to a fresh GYEP medium, a 15-ADON inducing medium (N-limiting), minimal medium (MM) containing ammonium chloride (MM NH4Cl), MM containing sodium nitrate (NaNO3), and MM containing no nitrogen (MM -N) and incubated for an additional 4 h. Relative expression (RQ) of FGSG_03881 (white), Tri10 (grey) and Tri6 (black) was quantified with β-tubulin as standard and normalized to the GYEP conditions. (b) The expression of FGSG_03881 is independent of Tri6. Expression levels of FGSG_03881 (RQ) in the 15-ADON inducing medium (N-limiting) for the wildtype (white), Δtri6 (grey) and Δfgsg_03881 (black) strains are shown. The expression was normalized to the GYEP conditions with β-tubulin as standard. The primers used in the qRT-PCR experiments are listed in . (c) HPLC analyses of the Δfgsg_03881 and wildtype strains in 15-ADON inducing medium compared with a 15-ADON toxin standard; quantification using peak integration and a standard curve determined that the cultures had 120 ppm and 166 ppm of 15-ADON, respectively. Absorbance values were offset to reduce overlap between samples. The result is representative of three independent biological experiments.
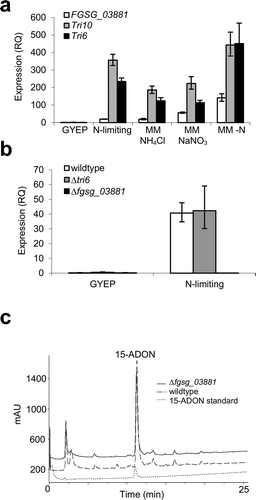
Fig. 4 Growth rate of F. graminearum is dependent on nitrogen availability. (a) PCR confirmation of pFGSG_03881::GFP transcriptional fusion strain. Genomic DNA was extracted from the wildtype (1) and pFGSG_03881::GFP (2) strains and amplified with primers specific to the Geneticin marker gene (Geneticin) and green fluorescent protein (GFP). The primers used are listed in . (b) The growth of the pFGSG_03881::GFP transcriptional fusion strain was measured in minimal media appended with different nitrogen sources at 2 g L−1. Growth was measured over a 5-day period at 620 nm in a 96-well plate in a POLARstar Optima plate reader. All data points are the average of three biological replicates and are presented as a percentage of the maximum observed value.
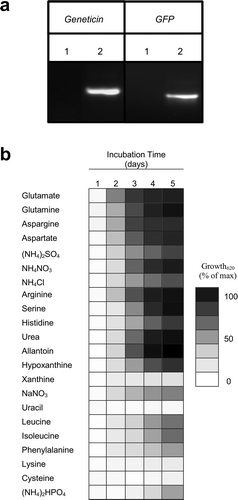
Fig. 5 Normalization of FGSG_03881 expression. The growth media containing glutamine, sodium nitrate and ammonium phosphate dibasic were selected as reference conditions for relative quantification. Before normalization: Fungal growth (620 nm) and GFP fluorescence (520 nm) were measured simultaneously. Both measurements are represented as transmittance units (×103) in the y-axis. After normalization: GFP fluorescence expressed as a function of growth for all three media conditions. A fourth order polynomial equation was derived from each of the normalized growth curves with coefficient of determination R2 glutamine = 0.99, = 0.99,
= 0.93.
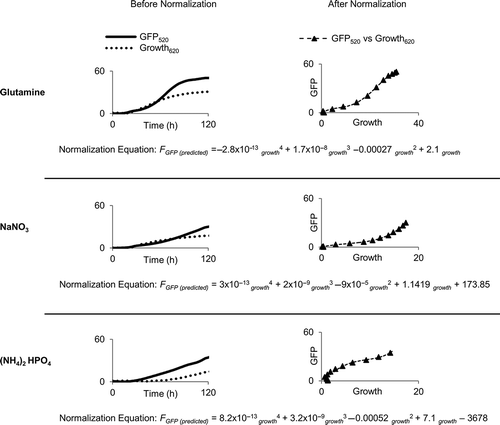
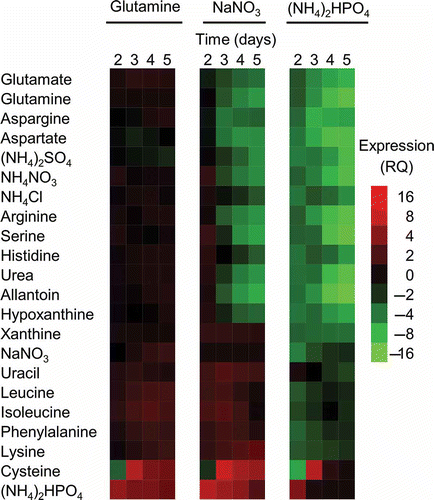
Fig. 6 (Colour online) Expression of FGSG_03881 in 22 different nitrogen conditions. The heat map shows relative expression (RQ) of FGSG_03881 determined using a ratio of the predicted GFP and measured GFP fluorescence values; predicted values were obtained using the formula derived from three different reference conditions (sodium nitrate, glutamine and ammonium phosphate dibasic) that are found in . The RQ is shown by the gradation scale with high expression represented by red and low expression represented by green. This result is a representative of three independent biological replicates.