Figures & data
Fig. 1 (Colour online) Geographical distribution of the visual survey locations for peach yellows disease in Shaanxi. The symbol (+) indicates where the disease occurred and (-) for the negative locations.
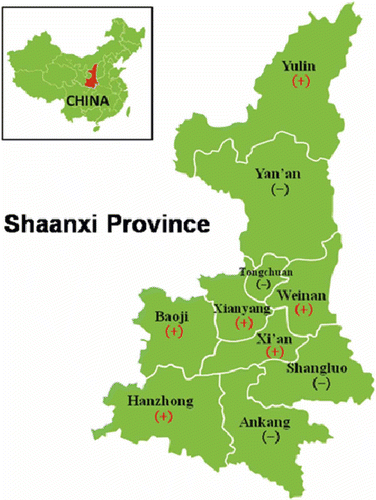
Fig. 2 Symptoms in peach trees infected with phytoplasmas. The diseased peach trees exhibited yellows and dieback (A, B), small and wilted fruit (C) and tattered appearance of leaves (D).
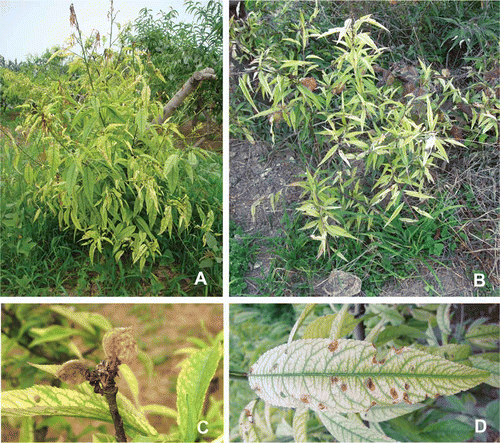
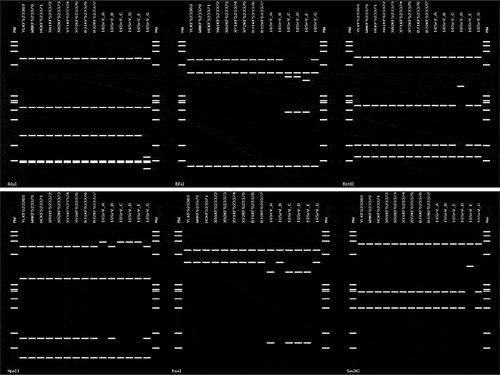
Table 1. 16SrRNA gene sequences of phytoplasmas used for the phylogenetic analysis.
Table 2. Results of nested PCR detection for phytoplasma in peach.
Table 3. Identities between the nine sequences determined in this study and its homogeneous sequences.
Fig. 3 Transmission electron micrographs of an ultrathin section from a symptomatic sample (A) and a healthy sample (B). The black arrows indicate some phytoplasma bodies.
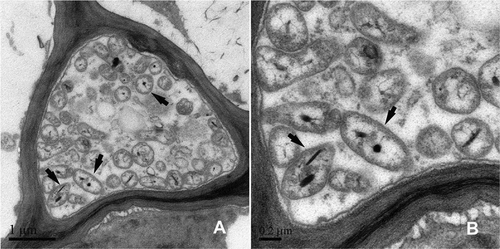
Fig. 4 Phylogenetic tree inferred from the 49 16S rRNA gene sequences of phytoplasmas using MEGA5. Acholeplasma laidlawii was used as an out-group reference.
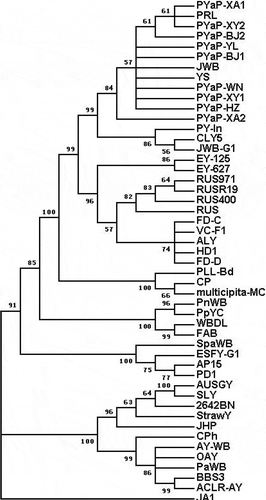
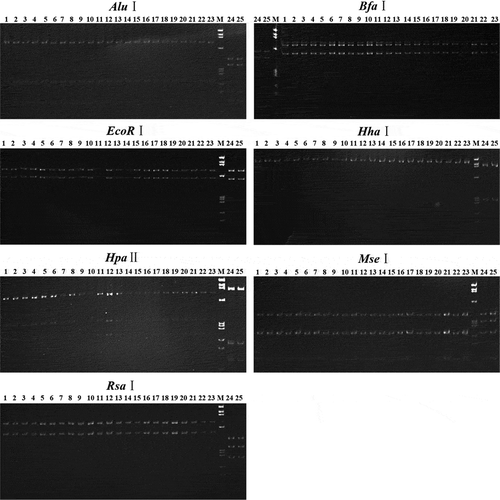
Fig. 5 Separation of nested PCR products after digesting with enzymes. Lanes 1–21, PYaP. Lane 22, graft-transmitted PYaP. Lane 23, JWB. Lanes 24, CWB. Lane 25, MD. Lane M, DNA marker ΦX174 HaeIII digest DNA ladder; band sizes from top to bottom are 1353, 1078, 872, 603, 310, 281, 271, 234, 194 and 118 bp.