Figures & data
Fig. 1 Construction of the knockout and overexpression vectors. a, The pCAMBIA1301B knockout vector. b, The fragment of the EcoRI-left arm-KpnI-PtrpC-bar-TtrpC-BamHI-right arm-XbaI containing the selectable marker bar gene coding for the resistance of glufosinate and four restriction sites (EcoRI, KpnI, BamHI and XbaI). c, The fragment of the HindIII-PtrpC-CBX-TtrpC-AhdI containing a selectable marker CBX gene coding for carboxin resistance and containing two restriction sites (HindIII and AhdI). d, The fragment of the Zn(II)2Cys6 transcription factor gene (rolP) containing two restriction sites (EcoRI and XbaI).
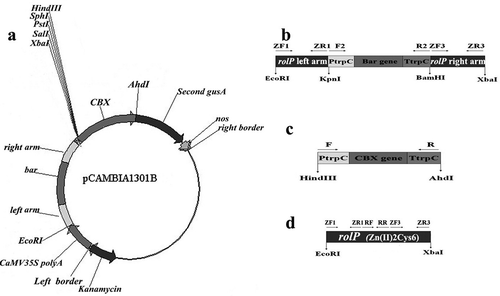
Table 1. Primers used for the reconstruction of the binary vector.
Fig. 2 Molecular analysis of rolP. a, PCR products of the rolP gene of the R147 strain by the Tail-PCR method. b, Phylogenetic analysis of rolP and its homologues from other fungal species.
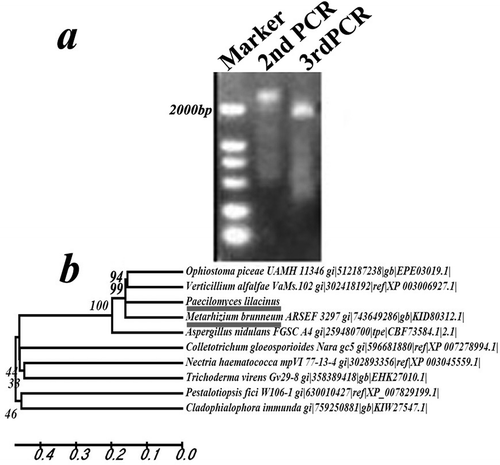
Fig. 3 Validation of the rolP knockout and overexpression strains. a, PCR analyses of WT and transformants. Lane 1, Takara 250 bp DNA ladder; Lanes 2–4, amplification of rolP. 2, the knockout strain ΔrolP, 3, the overexpression strain Ov-Pl36-1, 4, the wild type strain Pl36-1. Lanes 5–6, amplification of the CBX resistance gene. 5, the Ov-Pl36-1 strain and 6, the WT strain. b, Southern blot analysis of the rolP gene in the colonies of WT and transformants. Lane 1: The pCAMBIA1301B vector; Lane 2, the ΔrolP strain; Lane 3, the Ov-Pl36-1 strain; Lane 4, the WT strain. c, Southern blot analysis of the CBX gene in the colonies of WT and transformants. Lane 1, the pCAMBIA1301B vector; Lane 2, the Δrolp strain; Lanes 3, the Ov-Pl36-1 strain; Lane 4, the WT strain.
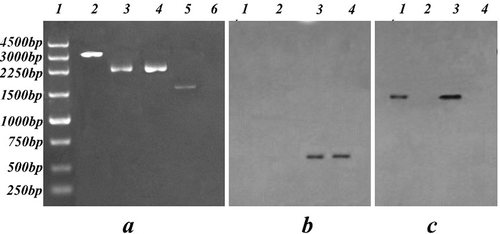
Table 2. Radial growth and sporulation yield of P. lilacinus strains.
Fig. 4 (Colour online) Determination of leucinostatins from P. lilacinus strains. Leucinostatin production of P. lilacinus 36-1 and its transformants was determined by MALDI-TOF MS. Leucinostatin A molecular weight: 1218. Leucinostatin B molecular weight: 1204. a, The metabolites of the WT strain P. lilacinus 36-1. b, The metabolites of the knockout strain ΔrolP. c, The metabolites of the overexpressed strain Ov-Pl36-1.
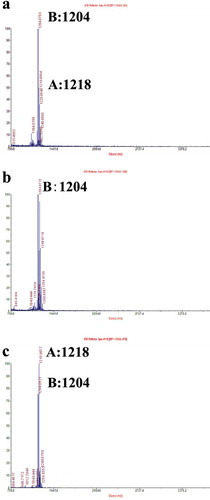
Fig. 5 Pathogenicity test of P. lilacinus strains against M. incognita. The pathogenicity of the P. lilacinus strains was determined by examining the effectiveness of each strain against eggs and second-stage juveniles (J2) of M. incognita. a, The results of corrected-parasitized eggs. b, The results of the J2-corrected mortality rate.
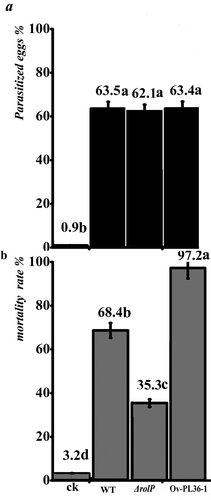
Fig. 6 Semi-quantitative reverse transcription PCR analysis of rolP genes in P. lilacinus. a, The expression levels of rolP in the WT, ΔrolP and Ov-pl36-1 strains at 48 h. b, The expression levels of rolP in the WT strain at different infection periods.
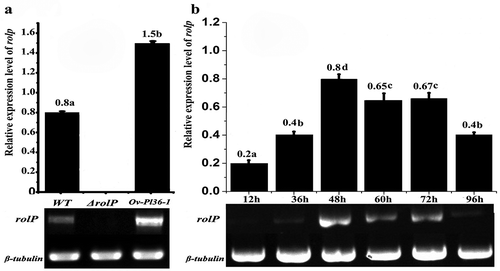
Fig. 7 Effect of carbon and nitrogen sources on WT toxicity and rolP gene expression. The WT strain was cultured in medium B supplemented with various carbon and nitrogen sources. The toxicity of the cell-free culture was examined against root-knot J2. SqRT-PCR was used to examine the mRNA level of the rolP gene. a, b and c show medium B with carbon sources. d, e and f show medium B with nitrogen sources. The mortality rates of J2 are shown in (a) and (d). The relative expression levels of the rolP gene are shown in (b) and (e). The initial and final pH values are shown in (c) and (f). The data shown here are the means ± SD (n = 3). *, P ≤ 0.05.
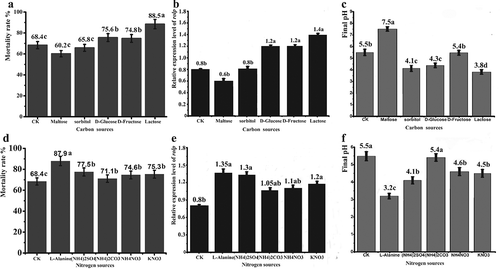
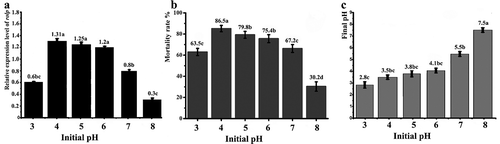
Fig. 8 Effect of pH on WT toxicity and the rolP gene. The WT strain was cultured in medium B with different pH values ranging from3.0 to 8.0. a, The mRNA level of the rolP gene. b, The mortality rates of root-knot J2 in cell-free culture. c, Final pH values of cell-free culture. The data shown here are the means ± SD (n = 3). *, P ≤ 0.05.