Figures & data
Table 1. Classification of 28 phytoplasma strains based on 16S rDNA, ribosomal protein and secY sequences into RFLP subgroups.
Fig. 1 RFLP analysis of 16S rDNA, rp operon and secY sequences of phytoplasma strains. a, RFLP profiles of the 16S rDNA nested PCR products amplified with primer pairs R16F2n/R2 and digested using restriction endonucleases BfaI, HhaI and MseI. b, RFLP profiles of the rp operon sequences (containing rpl22 and rps3 genes) amplified with primer pairs rPF1/rpR1 digested using restriction endonucleases AluI, Tsp5091 and MseI. c, RFLP profiles of the secY gene sequences amplified with the primer pairs AYsecYF1/AYSecYR1 using restriction endonucleases AluI, Tsp5091 and MseI. Lanes M is GeneRuler 1 kb plus DNA ladder (Life Technologies) molecular weight marker. Digests were separated on 3% super fine resolution agarose gel (SFR) except for MseI and Tsp5091 digests of rp and secY genes which were separated on 5% Agarose SFR gel. Samples Bn03 and Bn06: subgroup IA; Bn10 and Bn16: subgroup IB; Bn11 and Bn24: mixed infection.
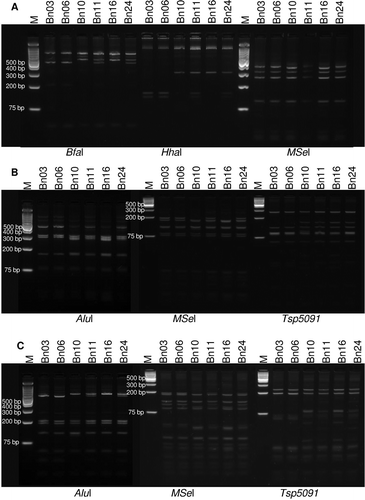
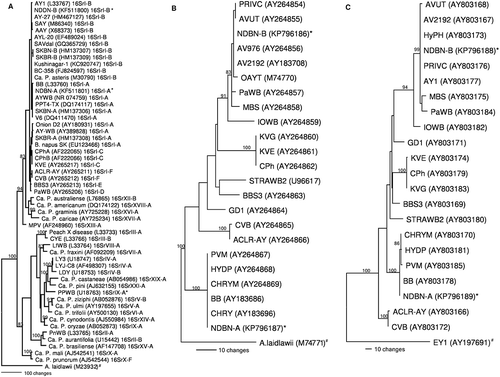
Fig. 2 Phylogenetic trees constructed by parsimony analyses based on three genetic regions. a, 16S rDNA (using representative sequences of several groups of phytoplasmas) b, rp operon (using representative sequences of the rp operon from AY subgroups) and c, secY (using representative sequences of the secY gene from AY subgroups) sequences using PAUP 4.0 version (Swofford Citation2002). Reference strains for 16S rDNA (Olivier et al. Citation2010), rp operon (Lee et al. Citation2004), and secY (Lee et al. Citation2006) were as described in the original papers. Accessions with an * represent the strains sequenced in this study and strains with # are used as outgroups. Trees were constructed via step-wise addition by 100 replicates of a heuristic search employing tree bisection and reconnection algorithm to find optimal trees. Branch lengths are proportional to the number of inferred character state transformations. Bootstrap analyses (1000 replicates) were conducted to determine the stability and support for the inferred clades. Bootstrap values above 80% are shown on main branches.