Figures & data
Table 1. Putative housekeeping genes, location on the array, code and oligonucleotide sequences evaluated in this study.
Fig. 1 Layout of multilocus oligonucleotides spotted on array. Uni506_16S was a positive control used to normalize hybridization intensity; NS, not spotted, negative control. 1, 16S rRNA; 2, leuS, 3, cpsD; 4, rpoB; 5, gyrB; 6, rpoB (b6 & c6) and leuS (d6). The corresponding sequences of all oligonucleotides are listed in .
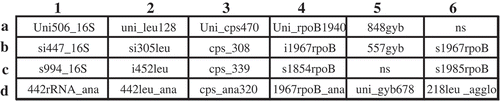
Fig. 2 Maximum likelihood inferred phylogenetic tree of Pantoea species from gyrB-leuS-rpoB concatenated sequences. Pantoea stewartii subsp. stewartii strains seem to be homogeneous while those of P. stewartii subsp. indologenes are heterogeneous. General time reversible substitution model was used with 1000 bootstrap replicates. Tatumella spp. used as outgroup.
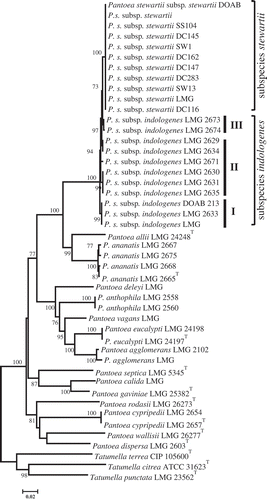
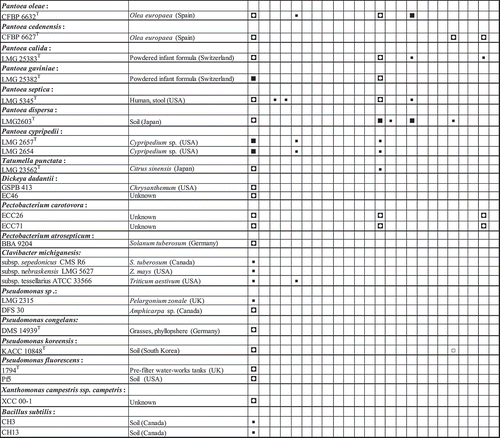
Fig. 3 Summary of hybridization patterns of multiplex digoxigenin-labelled PCR amplicons of Pantoea stewartii and other bacterial strains to an array of subspecies-, species- and group-specific oligonucleotides on nylon membranes. Chemiluminograms were scanned at 600 dpi using HP ScanJet 5590 scanner, and grey scale values of each dark spot, computed with ImageJ software and normalized using oligonucleotide Uni506_16S. Normalized signals are indicated by the following symbols: □, <0.1 (not detected); ▪, 0.15–0.39; ◘, 0.40–0.79; ■, >0.80. The locations of the oligonucleotides given at the top correspond to their locations on the membranes in the array. Oligonucleotides are presented in . The type strains were obtained from BCCM/LMG bacterial collection, Belgium or the Collection of Plant associated Bacteria (CFBP), France. Strains with prefixes SW or DC were kindly provided by Dr David Coplin.
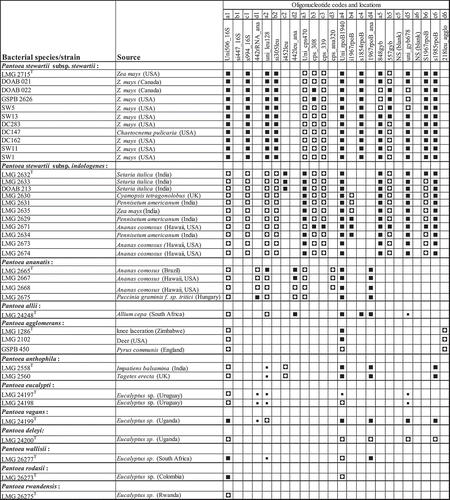
Fig. 4 Hybridization profiles of 18 Pantoea species and subspecies, Pectobacterium and Dickeya (former Erwinia members) and Tatumella punctata. Note specificity of detection of Pantoea stewartii subsp. stewartii LMG 2715T and differentiation from Pantoea stewartii subsp. indologenes based on oligonucleotide s1854rpoB (location c4).
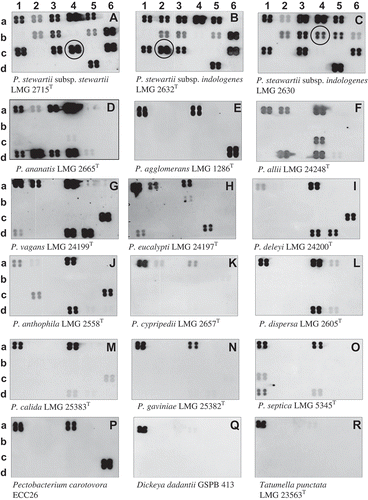
Fig. 5 Chemiluminograms showing specific detection of Pantoea stewartii subsp. stewartii DOAB 021 on corn plants infected or not under growth chamber conditions. a, non-infected plant, and b, infected plant. Leaf lesions (40 mg) were processed as described in Tambong et al. (Citation2008). No DNA extraction was performed. Four plants per treatments were analysed with consistent hybridization pattern obtained. Row numbers represent the different genes used: 1, 16S rRNA; 2, leuS; 3, cps; 4, rpoB; 5, gyrB; 6, mix: rpoB (b6 & c6) and leuS (d6) spotted on the membrane-based array.
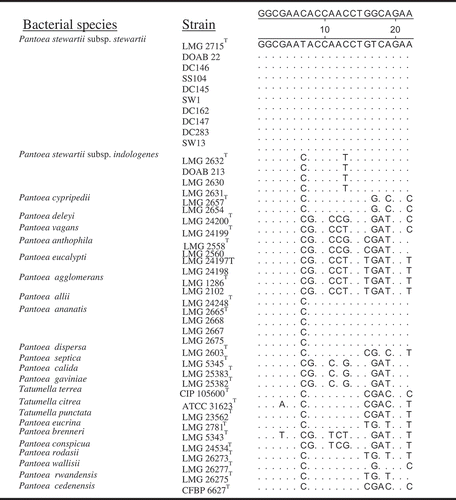
Fig. 6 Alignment of rpoB-derived oligonucleotide (s1854rpoB) sequences showing nucleotide mismatches and specificity to P. stewartii subsp. stewartii compared with other strains of Pantoea and Tatumella. Note nucleotide mismatches of oligonucleotide s1854rpoB are almost at the centre for effective differentiation.