Figures & data
Table 1. ASPV incidence on apple and pear trees surveyed in this study in different areas.
Table 2. Genetic distance between and within groups clustered in phylogenetic trees based on ASPV CP and TGB sequences.
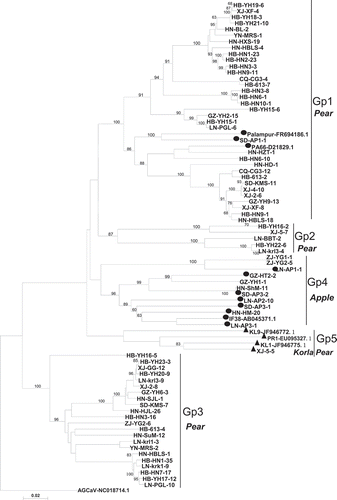
Fig. 1 Phylogenetic trees of complete CP (a) and TGB (b) sequences of ASPV isolates in this study and global isolates available in GenBank. Reference isolates are named according to their isolate name with GenBank accession numbers. Isolates reported herein are indicated by isolate abbreviations and number followed by the sequenced CP or TGB clone number. Sequences isolated from apple are indicated with the symbol ●, whereas sequences from Korla pear are denoted with the symbol ▲ and all other sequences are from pear isolates. The tree was constructed by the neighbour joining method implemented by MEGA6. Bootstrap analysis with 1000 replicates was performed. Only ≥50% bootstrap values are shown, and branch lengths are proportional to the genetic distances. Bar, 0.05 substitutions per site.
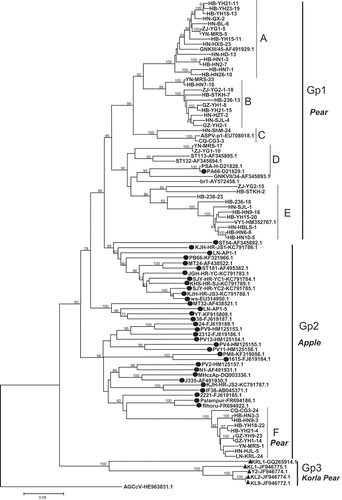
Table 3. List of putative recombination events involving ASPV CP and TGB sequences.
Table 4. Analysing the selection pressures (dN/dS) of different ASPV genes.
Table 5. Population genetic parameters and neutrality tests calculated for ASPV CP and TGB based on geographic origins or variant groups.
Fig. 2 Multiple alignments of amino acids of 16 representative ASPV CP sequences. 2–3 sequences from each subgroup in the phylogenetic tree in were selected. LN-AP1-1 (arrow) from an apple isolate was used as a reference sequence. The deleted regions are represented by rectangular boxes and the inserted regions are represented by oval boxes.