Figures & data
Fig. 1 (Colour online) External and internal necrosis in potatoes ‘Chieftain’ collected in a field in New Brunswick, Canada. Photos were taken 3 months after harvesting.

Table 1. Summary of genomic RNA sequences of Potato mop-top virus (PMTV) isolates in Canada and China as well as other PMTV isolates available in databases.
Table 2. Nucleotide (shaded) and amino acid (non-shaded) sequence identities between different isolates of Potato mop-top virus in China and Canada.
Fig. 2 (Colour online) (a) Bayesian phylogenetic tree of the genomic RNA1 of Potato mop-top virus (PMTV) isolates. Phylogenetic analyses were carried out with the Bayesian inference (BI) in MrBayes v3.22. For each node, the Bayesian posterior probabilities are given above branches (only shown >50%) and isolate grouping based on genetic similarity and nodes with posterior probabilities greater than 70% are indicated. The nucleotide sequence accession number of each isolate is shown in parentheses. The newly sequenced isolates are indicated by ‘*’. The distance unit is substitutions/site. (b) Analysis of the sequence identities between ‘Guangdong’-RNA 1 (query sequence) and RNA1 of other isolates (reference sequences) using SimPlot. The similarities (y-axis, 0.9–1) were plotted along the nucleotide sequence (x-axis, nt 1–6000). The window covered 500 nucleotides and moved along the alignment with 20 nucleotides with every step. Other parameter settings were: distance model, Kimura (2-parameter); tree model, neighbour-joining; bootstrap replicates, 100; parental threshold, 70. Nucleotide sequence accession numbers for isolates are the same as shown in (a).
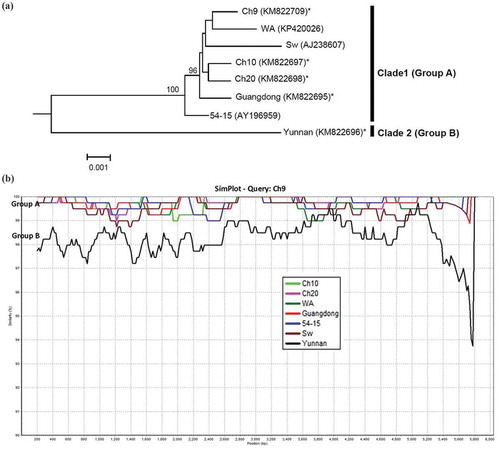
Fig. 3 (Colour online) (a) Bayesian phylogenetic tree of the genomic RNA2 of Potato mop-top virus (PMTV) isolates. Phylogenetic analyses were carried out with the Bayesian inference (BI) in MrBayes v3.22. For each node, the Bayesian posterior probabilities are given above branches (only shown >50%) and isolate grouping based on genetic similarity and nodes with posterior probabilities greater than 70% are indicated. The nucleotide sequence accession number of each isolate is shown in parentheses. The newly sequenced isolates are indicated by ‘*’. The distance unit is substitutions/site. (b) Analysis of the sequence identities between ‘Guangdong’-RNA2 (query sequence) and RNA2 of other isolates (reference sequences) using SimPlot. The similarities (y-axis, 0.9–1) were plotted along the nucleotide sequence (x-axis, 0–3,100). The window covered 500 nucleotides and moved along the alignment with 20 nucleotides with every step. Other parameter settings were: distance model, Kimura (2-parameter); tree model, neighbour-joining; bootstrap replicates, 100; parental threshold, 70. Nucleotide sequence accession numbers are the same as shown in (a).
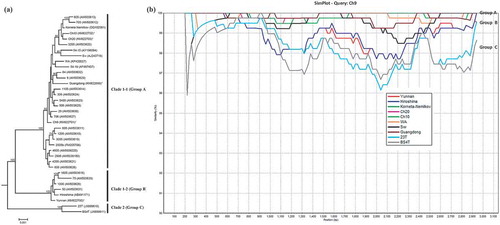
Fig. 4 (Colour online) (a) Bayesian phylogenetic tree of the genomic RNA3 of Potato mop-top virus (PMTV) isolates. Phylogenetic analyses were carried out with the Bayesian inference (BI) in MrBayes v3.22. For each node, the Bayesian posterior probabilities are given above branches (only shown >50%) and isolate grouping based on genetic similarity and nodes with posterior probabilities greater than 70% are indicated. The nucleotide sequence accession number of each isolate is shown in parentheses. The newly sequenced isolates are indicated by ‘*’. The distance unit is substitutions/site. (b) Analysis of the sequence identities between RNA3 of ‘Guangdong’ (query sequence) and RNA3 of other isolates (reference sequences) using SimPlot. The similarities (y-axis, 0.9–1) were plotted along the nucleotide sequence (x-axis, 0–2900). The window covered 500 nucleotides and moved along the alignment with 20 nucleotides with every step. Other parameter settings were: distance model, Kimura (2-parameter); tree model, neighbour-joining; bootstrap replicates, 100; parental threshold, 70. Nucleotide sequence accession numbers for isolates are the same as shown in (a).
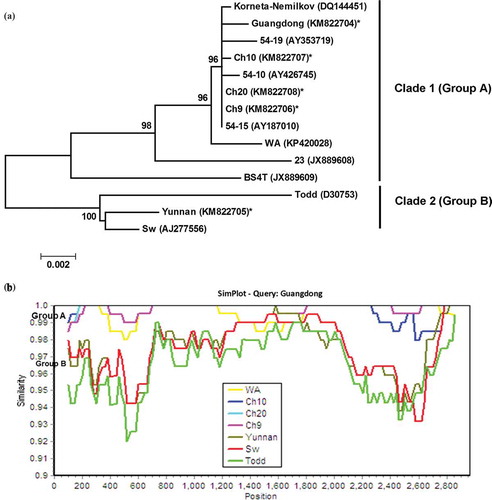
Fig. 5 RT-PCR differentiation of RNA3 belonging to different phylogenetic groups in Potato mop-top virus (PMTV). (a) Alignment of sequences of RNA3 of PMTV for designing primers for detection and differentiation of RNA3 belonging to phylogenetic Group A (KP420028_Wa, CA-Ch10, CA-Ch9, CA-Ch20, Guangdong, AY187010_54–15) and Group B (D30753_Todd, Yunnan and AJ277556_Sw). Primers F459 and R920 are group-non-specific whereas primer F588-YN is group-B specific. (b) Schematic diagram of PCR amplification products for differentiation of PMTV-RNA3 belonging to phylogenetic Groups A and B. Target segments and resulting PCR products are illustrated. (c) RT-PCR assay results for differentiating RNA3 phylogenic groups. Left panel, simplex RT-PCR with primers F459 and R920; central panel, simplex RT-PCR with primers F588-YN and R920; right panel, duplex RT-PCR with F459, F588-YN and R920. Lanes 1–2, PMTV isolate ‘Guangdong’; lanes 3–4: PMTV isolate ‘Yunnan’; lane ‘-’, negative control; lane M = DNA ladder.
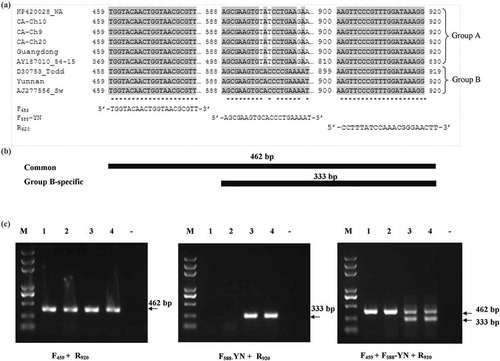
Table 3. Genomic RNA type of Potato mop-top virus based on the phylogenetic grouping and sequence similarity scanning.
